Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Elena A. Pudova | -- | 2214 | 2023-12-19 13:15:43 | | | |
| 2 | Rita Xu | Meta information modification | 2214 | 2023-12-20 02:39:26 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Pudova, E.; Kobelyatskaya, A.; Emelyanova, M.; Snezhkina, A.; Fedorova, M.; Pavlov, V.; Guvatova, Z.; Dalina, A.; Kudryavtseva, A. Non-Coding RNAs in Prostate Cancer. Encyclopedia. Available online: https://encyclopedia.pub/entry/52928 (accessed on 07 February 2026).
Pudova E, Kobelyatskaya A, Emelyanova M, Snezhkina A, Fedorova M, Pavlov V, et al. Non-Coding RNAs in Prostate Cancer. Encyclopedia. Available at: https://encyclopedia.pub/entry/52928. Accessed February 07, 2026.
Pudova, Elena, Anastasiya Kobelyatskaya, Marina Emelyanova, Anastasiya Snezhkina, Maria Fedorova, Vladislav Pavlov, Zulfiya Guvatova, Alexandra Dalina, Anna Kudryavtseva. "Non-Coding RNAs in Prostate Cancer" Encyclopedia, https://encyclopedia.pub/entry/52928 (accessed February 07, 2026).
Pudova, E., Kobelyatskaya, A., Emelyanova, M., Snezhkina, A., Fedorova, M., Pavlov, V., Guvatova, Z., Dalina, A., & Kudryavtseva, A. (2023, December 19). Non-Coding RNAs in Prostate Cancer. In Encyclopedia. https://encyclopedia.pub/entry/52928
Pudova, Elena, et al. "Non-Coding RNAs in Prostate Cancer." Encyclopedia. Web. 19 December, 2023.
Copy Citation
Chemotherapy based on taxane-class drugs is the gold standard for treating advanced stages of various oncological diseases. Drug resistance is the result of a combination of different events in the tumor cells under the influence of the drug, a comprehensive understanding of which has yet to be determined.
prostate cancer
chemoresistance
docetaxel
microRNAs
lncRNAs
circRNAs
1. Introduction
Prostate cancer (PCa) is a socially significant cancer and is the second most common type of cancer in men worldwide [1]. Most patients with PCa have localized disease and are treated with radical prostatectomy and/or radiation therapy followed by androgen deprivation therapy. However, within 10 years after androgen deprivation in patients, in 10–20% of cases, the disease develops into a prognostically unfavorable form—castration-resistant prostate cancer (CRPC), which is characterized by a significant deterioration in the quality of life and high mortality of patients. The median overall survival of patients with CRPC is less than 2 years [2]. Chemotherapy based on taxane-class drugs, such as paclitaxel and docetaxel, is the gold standard for the first-line therapy in this patient category. Docetaxel is the most commonly used first-line chemotherapy drug for CRPC. However, despite its widespread use in therapy, most patients eventually develop resistance, which is one of the reasons for the ineffectiveness of chemotherapy in patients [3].
Drug resistance is conventionally divided into two main classes: primary (existing initially) and acquired [4]. Primary resistance is characterized by the presence of various factors in tumor cells prior to the action of the drug, whereas acquired resistance represents a stepwise and slower process involving various molecular genetic and epigenetic events in the presence of the drug [5]. The mechanisms underlying acquired drug resistance are quite complex and involve alterations in the regulation of numerous genes and different signaling pathways, which act independently or in combination with other factors to inhibit the function of taxanes in tumor cells. Several studies have identified a range of mechanisms involved in the development of taxane resistance, yet the full extent of the picture is still to be determined [6].
For a long time, non-coding RNAs were considered as by-products of transcription with little biological significance, in contrast to messenger RNAs (mRNAs). However, since the development of approaches such as high-throughput sequencing and in-depth bioinformatic analyses, new types of RNAs that do not encode proteins, collectively known as non-coding RNAs (ncRNAs), have been discovered. Currently, the most studied types of “classic ncRNAs” are microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). It has been shown that these molecules participate in many signaling cascades, regulate various physiological processes, and also play a role in diseases. Moreover, many ncRNAs have been identified as tumor suppressors/oncogenic factors in various types of cancer, including prostate cancer, and they may also participate in the development of drug resistance [7].
2. MicroRNAs
MicroRNAs (miRNAs) are regulatory molecules with a length of 20–22 nucleotides that are formed from longer stem-loop structures and can bind to and inhibit mRNA. These molecules are transcribed as primary miRNAs (pri-miRNAs) and processed in the nucleus by the proteins Drosha and Dgcr8 into precursor miRNAs (pre-miRNAs). After export to the cytoplasm, pre-miRNAs are cleaved, forming a miRNA/miRNA duplex. Only one of the two miRNAs formed will exert its inhibitory function, while the other undergoes degradation. The total number of known human miRNAs is constantly expanding and currently includes 1917 precursors and 2654 mature molecules (miRBase, version 22.1).
Many miRNAs are often suppressed in drug-resistant cells, but under normal cellular conditions, they help regulate signaling pathways that promote cell survival. The ability to avoid programmed cell death, apoptosis, is one of the key characteristics of tumor cells that ensures their survival. Tumor cells, like normal cells, also possess proteins from the BCL-2 family that regulate apoptosis. These include Bad and Bax, which initiate cascades that activate apoptosis, and Bcl-2, Bcl-XL, and Mcl-1, which inhibit the apoptosis process [6]. Several microRNAs (miRNAs) have been demonstrated through various studies to regulate the development of docetaxel resistance in PCa cells by enhancing cell survival and inhibiting apoptosis. These miRNAs include miR-143, the miR-200 family, miR-21, miR-129-5p, miR-27a, miR-27b, miR-34a, miR-183, miR-195, miR-223-3p, miR-323, and miR-4735-3p (Figure 1).
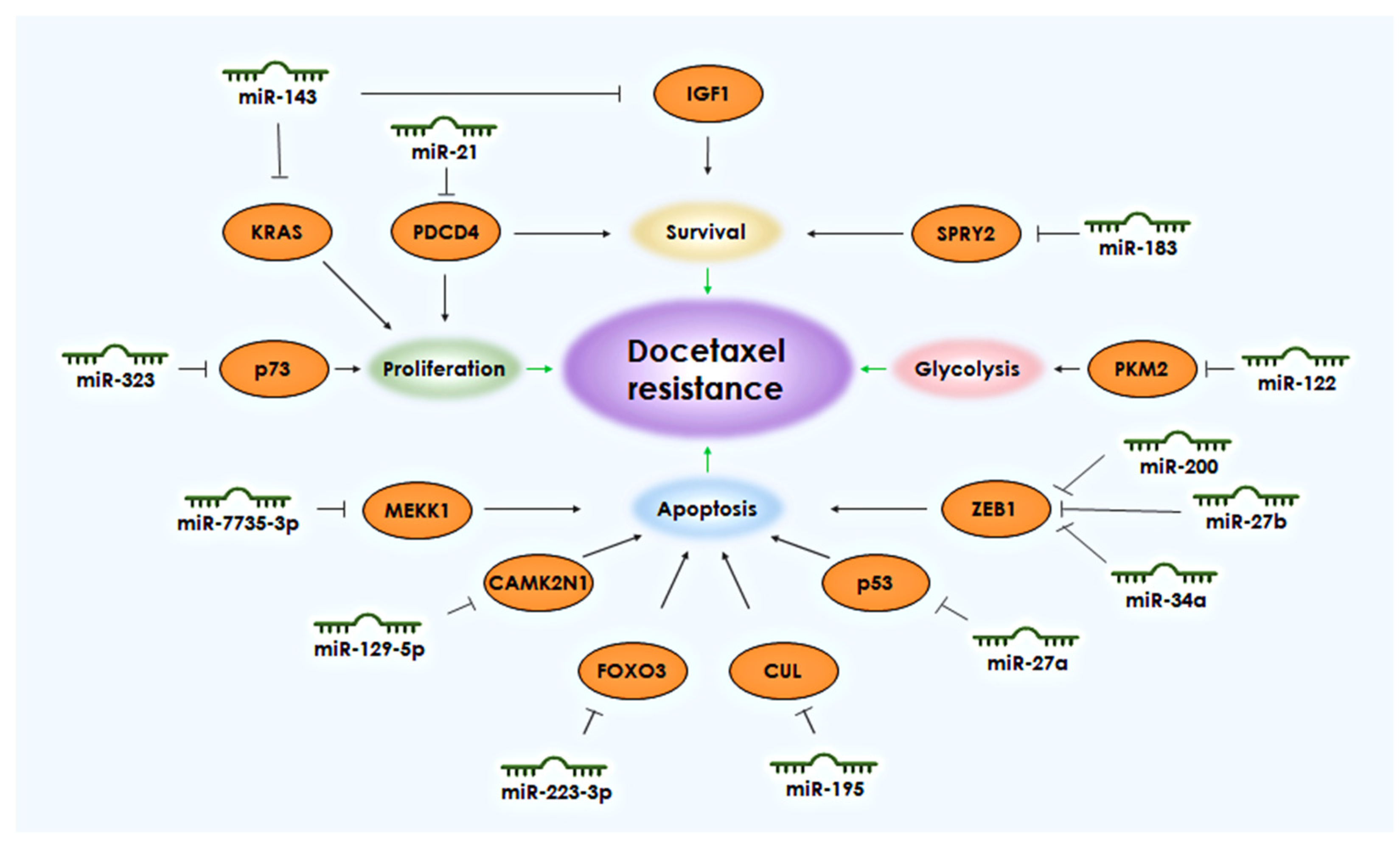
Figure 1. miRNAs and their targets associated with the development of resistance to docetaxel in PCa. The black arrows shows the connection between the target and the biological process. The green arrows indicate the connection of the process with the development of resistance to docetaxel.
The downregulation of miR-143 can induce docetaxel resistance by enhancing the regulation of insulin-like growth factor-1 receptor (IGF-1R) and insulin receptor substrate 1 (IRS1), resulting in the activation of downstream signaling molecules such as VEGF (vascular endothelial growth factor A). Activation of the IGF-1R/IRS1/VEGF signaling cascade promotes the survival pathways of PCa cells and reduces sensitivity to docetaxel [8]. Furthermore, it has been found that miR-143 can regulate the KRAS (KRAS Proto-Oncogene, GTPase) gene, which is involved in the activation of the oncogenic MAPK/Ras pathway, and the overexpression of miR-143 can increase the sensitivity of cells to docetaxel [9].
Regarding the expression of the miR-200 family of regulatory molecules, a correlation has been found with participants involved in epithelial–mesenchymal transition such as E-cadherin and ZEB1 (Zinc Finger E-Box Binding Homeobox 1). Decreased expression of miR-200 family members leads to increased expression of ZEB1, which negatively regulates E-cadherin. It was experimentally shown that the overexpression of miR-200 family members in PCa cells resulted in increased expression of E-cadherin and increased apoptosis induced by docetaxel [10]. Inhibition of epithelial–mesenchymal transition by suppressing the expression of the ZEB1 gene has also been found in the context of docetaxel resistance in PCa cells due to the overexpression of miR-27b and miR-34a [11].
Similarly, miRNAs whose expression is normally at low levels can excessively stimulate cell survival pathways or inhibit pro-apoptotic factors. For example, it has been shown that miR-21 can inhibit programmed cell death 4 (PDCD4), leading to a resistant phenotype in PCa cells. Inhibiting miR-21 in this case increases the expression of PDCD4 and restores sensitivity to docetaxel [12].
Regarding miR-129-5p, it has been shown that increased expression of this regulatory molecule leads to the suppression of the CAMK2N1 (Calcium/Calmodulin Dependent Protein Kinase II Inhibitor 1) gene, resulting in the inhibition of apoptosis in PCa cells and the development of docetaxel resistance [13].
The most well-known tumor suppressor gene is TP53 (Tumor Protein P53), and alterations in its expression or function are often associated with resistance to standard anti-cancer agents. In the case of PCa, it has been shown that exosomal miR-27a can induce resistance to cisplatin, docetaxel, and doxorubicin in recipient cells by degrading p53 mRNA, resulting in reduced expression of the negative regulator of the PI3k/Akt signaling pathway, PTEN (Phosphatase and Tensin Homolog). This leads to decreased dephosphorylation of PIP3, increased cell survival, and proliferation of tumor cells [14].
miR-183 also regulates cell survival pathways in PCa and contributes to docetaxel resistance. It has been shown that miR-183 regulates the expression of the tumor suppressor gene SPRY2 (Sprouty RTK Signaling Antagonist 2), and its activation inhibits SPRY2 expression, thereby promoting resistance to docetaxel [15].
The expression level of miR-195 is often decreased in docetaxel-resistant cells. It has been demonstrated that the CUL ( Cullin 3) gene, which is involved in the anti-apoptotic mechanism of docetaxel-resistant cells, is a direct target of miR-195. Thus, overexpression of miR-195 inhibits CUL gene expression and increases the sensitivity of PCa cells to docetaxel [16].
Regarding miR-223-3p, miR-323, and miR-4735-3p, their increased expression in PCa cells inhibits docetaxel-induced apoptosis. miR-223-3p directly targets the FOXO3 (Forkhead Box O3) gene, miR-323 targets the tumor suppressor p73, while miR-4735-3p inhibits the expression of the MEKK1 (Mitogen-Activated Protein Kinase Kinase Kinase 1; also known as MAP3K1) gene [17][18][19].
3. LncRNAs
Long non-coding RNAs (lncRNAs) are regulatory molecules that are more than 200 nucleotides long and can be transcribed from introns, exons, intergenic regions, or non-coding regions. These molecules can perform various functions, such as participating in transcription as factors, acting as “sponges” in protein–protein interactions, but their most interesting action lies in inhibiting miRNAs. Specific lncRNAs can act as competing endogenous RNAs (ceRNAs), which sequester or inhibit microRNAs involved in pro-apoptotic pathways [20]. The exact number of known lncRNA genes is constantly increasing, and currently, the number of registered lncRNAs has reached 100,000 [21]. There are now hundreds of thousands of cataloged lncRNAs and dozens of databases with carefully curated information, such as LncATLAS, LncBook, LNCipedia, lncRNAKB, and others [22].
The upregulation of key lncRNAs and the subsequent suppression of corresponding miRNAs is also associated with taxane resistance in PCa. Based on various studies, a number of lncRNAs have been identified whose interaction with miRNAs is associated with the development of docetaxel resistance in PCa: NEAT1, UCA1, PCAT1, DANCR, CASC2, MALAT1, and LINC01963 (Figure 2).
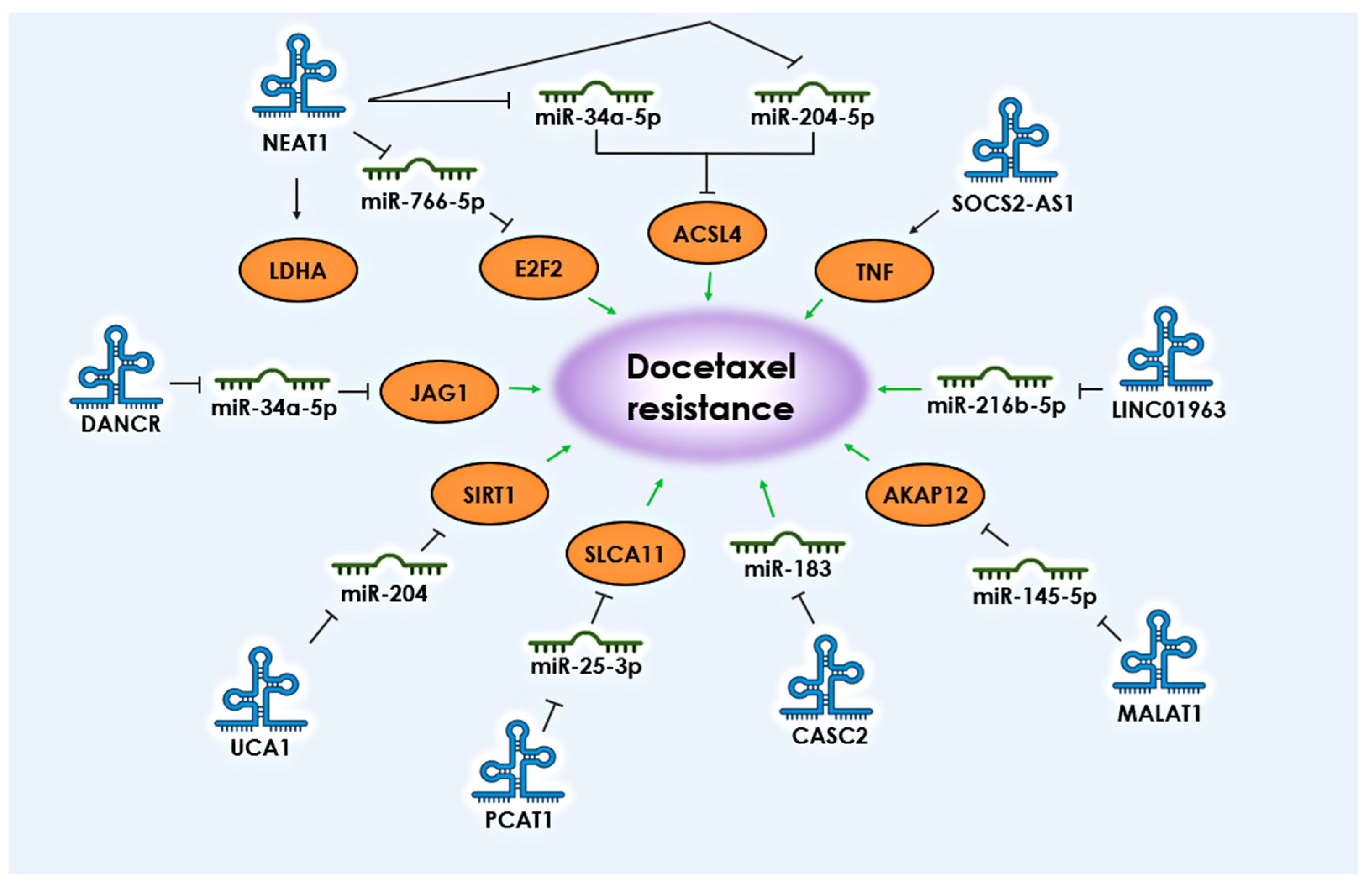
Figure 2. lncRNAs and their targets associated with the development of resistance to docetaxel in PCa. The black arrows shows the direction of communication between participants. Green arrows indicate the association with the development of docetaxel resistance.
According to research findings, lncRNA NEAT1 is considered a promising target for PCa treatment. Its increased expression leads to enhanced glycolysis by activating the LDHA (Lactate Dehydrogenase A) gene and suppressing T-cell immune surveillance [23]. Furthermore, NEAT1 plays an important role in PCa oncogenesis by acting as a sponge for miRNA-766-5p, resulting in increased expression of the transcription factor E2F3 [19]. High levels of NEAT1 expression are also associated with docetaxel resistance in PCa, through the suppression of miR-34a-5p and miR-204-5p, leading to increased expression of the ACSL4 (Acyl-CoA Synthetase Long Chain Family Member 4) gene, thereby promoting tumor progression and development of chemoresistance [24].
The activation of another lncRNA, UCA1, has also shown a connection with decreased expression of miR-204 and a positive correlation with increased expression of Sirt1, both of which regulate drug-induced apoptosis avoidance [25].
An important role in regulating the response to docetaxel in PCa has been found for lncRNA PCAT1, and there is increasing evidence of its involvement in PCa progression regulation [26]. It has been shown that the expression level of PCAT1 increases under the influence of TFAP2C (Transcription Factor AP-2 Gamma) in PCa cells. As a result of PCAT1 overexpression, the expression level of the SLCA11 (Solute Carrier Family 4 Member 11) gene is increased by binding to miR-25-3p. This process is crucial for preventing the death of tumor cells from ferroptosis and developing resistance to docetaxel [26].
LncRNA DANCR is significantly activated in docetaxel-resistant PCa. Research has shown that DANCR suppresses the degradation of JAG1 (Jagged Canonical Notch Ligand 1) induced by miR-34a-5p, thereby causing resistance to docetaxel [27]. Another lncRNA, CASC2, acts as a tumor suppressor in human malignancies, serving as a ceRNA for miR-183 and positively amplifying the expression of another tumor suppressor, SPRY2, a key antagonist of receptor tyrosine kinase signaling. The overexpression of CASC2 and SPRY2 can suppress the proliferation of PCa cells, promote their apoptosis, and increase sensitivity to docetaxel [28].
LncRNA MALAT1 is currently the most well-characterized lncRNA, and its aberrant expression is observed in various types of cancer, including PCa [29]. It has been shown that MALAT1 is involved in CRPC progression both in vivo and in vitro. Silencing MALAT1 can inhibit tumor cell proliferation by arresting the cell cycle at the G0/G1 phase [30]. Additionally, it has been demonstrated that MALAT1 enhances the expression of AKAP12 (A-Kinase Anchoring Protein 12) gene by directly targeting miR-145-5p, promoting resistance to docetaxel [31].
4. CircRNAs
Circular RNAs (circRNAs) represent a new class of non-coding single-stranded RNA molecules that are covalently linked to form a continuous closed loop and participate in the regulation of transcriptional and post-transcriptional gene expression [32]. Circular RNAs perform numerous unique and important biological functions: they act as traps for miRNAs or proteins, serve as scaffolds for circRNA–protein complexes, and recruit proteins to specific loci [32][33]. Additionally, some circRNAs can be translated into small unique peptides [34]. Research has shown that circRNAs promote tumor progression in various types of cancer by acting as RNA sponges and interacting with miRNAs, thereby enhancing gene expression [35].
CircRNAs are formed in circular transcripts through back-splicing of premature mRNAs, resulting in various types of circRNAs, such as exonic–intronic circRNAs (ElcircRNAs) (consisting of both exons and introns), circular intronic RNAs (formed by introns), exonic circRNAs (resulting from splicing of introns), and tRNA intronic circRNAs (formed by pre-tRNA splicing) [36]. To date, the total number of known circRNAs varies depending on the database. For example, the isoCirc database includes 107,147 full-length circRNA isoforms in 12 human tissues and one human cell line (HEK293), including 40,628 isoforms ≥ 500 nt in length [37]. The CircAtlas database contains 1,007,087 highly reliable circRNAs, with over 81.3% of them assembled into full-length sequences [38].
Based on the research, the crucial role of circRNAs in the development of chemoresistance in PCa has been highlighted, particularly regarding the molecules hsa_circ_0000735 and circFOXO3. The circular RNA hsa_circ_0000735 is activated in tissues and cells of docetaxel-resistant PCa. Functional analyses have shown that the suppression of hsa_circ_0000735 inhibits docetaxel resistance and suppresses tumor progression. Moreover, hsa_circ_0000735 can act as a sponge for miR-7, whose expression is decreased in docetaxel-resistant cells, thus promoting chemoresistance in PCa [39]. CircRNA circFOXO3 is one of the most studied circRNAs, and its inhibition has been shown to inhibit PCa progression and enhance docetaxel sensitivity by upregulating FOXO3 expression and repressing epithelial–mesenchymal transition of tumor cells [40].
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48.
- Mansinho, A.; Macedo, D.; Fernandes, I.; Costa, L. Castration-Resistant Prostate Cancer: Mechanisms, Targets and Treatment. Adv. Exp. Med. Biol. 2018, 1096, 117–133.
- Bumbaca, B.; Li, W. Taxane resistance in castration-resistant prostate cancer: Mechanisms and therapeutic strategies. Acta Pharm. Sin. B 2018, 8, 518–529.
- Velaei, K.; Samadi, N.; Barazvan, B.; Soleimani Rad, J. Tumor microenvironment-mediated chemoresistance in breast cancer. Breast 2016, 30, 92–100.
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348.
- Maloney, S.M.; Hoover, C.A.; Morejon-Lasso, L.V.; Prosperi, J.R. Mechanisms of Taxane Resistance. Cancers 2020, 12, 3323.
- Pavet, V.; Portal, M.M.; Moulin, J.C.; Herbrecht, R.; Gronemeyer, H. Towards novel paradigms for cancer therapy. Oncogene 2011, 30, 1–20.
- Niu, X.B.; Fu, G.B.; Wang, L.; Ge, X.; Liu, W.T.; Wen, Y.Y.; Sun, H.R.; Liu, L.Z.; Wang, Z.J.; Jiang, B.H. Insulin-like growth factor-I induces chemoresistence to docetaxel by inhibiting miR-143 in human prostate cancer. Oncotarget 2017, 8, 107157–107166.
- Xu, B.; Niu, X.; Zhang, X.; Tao, J.; Wu, D.; Wang, Z.; Li, P.; Zhang, W.; Wu, H.; Feng, N.; et al. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol. Cell Biochem. 2011, 350, 207–213.
- Puhr, M.; Hoefer, J.; Schafer, G.; Erb, H.H.; Oh, S.J.; Klocker, H.; Heidegger, I.; Neuwirt, H.; Culig, Z. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. Am. J. Pathol. 2012, 181, 2188–2201.
- Zhang, G.; Tian, X.; Li, Y.; Wang, Z.; Li, X.; Zhu, C. miR-27b and miR-34a enhance docetaxel sensitivity of prostate cancer cells through inhibiting epithelial-to-mesenchymal transition by targeting ZEB1. Biomed. Pharmacother. 2018, 97, 736–744.
- Shi, G.H.; Ye, D.W.; Yao, X.D.; Zhang, S.L.; Dai, B.; Zhang, H.L.; Shen, Y.J.; Zhu, Y.; Zhu, Y.P.; Xiao, W.J.; et al. Involvement of microRNA-21 in mediating chemo-resistance to docetaxel in androgen-independent prostate cancer PC3 cells. Acta Pharmacol. Sin. 2010, 31, 867–873.
- Wu, C.; Miao, C.; Tang, Q.; Zhou, X.; Xi, P.; Chang, P.; Hua, L.; Ni, H. MiR-129-5p promotes docetaxel resistance in prostate cancer by down-regulating CAMK2N1 expression. J. Cell. Mol. Med. 2020, 24, 2098–2108.
- Cao, Z.; Xu, L.; Zhao, S. Exosome-derived miR-27a produced by PSC-27 cells contributes to prostate cancer chemoresistance through p53. Biochem. Biophys. Res. Commun. 2019, 515, 345–351.
- Gao, W.; Lin, S.; Cheng, C.; Zhu, A.; Hu, Y.; Shi, Z.; Zhang, X.; Hong, Z. Long non-coding RNA CASC2 regulates Sprouty2 via functioning as a competing endogenous RNA for miR-183 to modulate the sensitivity of prostate cancer cells to docetaxel. Arch. Biochem. Biophys. 2019, 665, 69–78.
- Ma, X.; Zou, L.; Li, X.; Chen, Z.; Lin, Q.; Wu, X. MicroRNA-195 regulates docetaxel resistance by targeting clusterin in prostate cancer. Biomed. Pharmacother. 2018, 99, 445–450.
- Feng, Q.; He, P.; Wang, Y. MicroRNA-223-3p regulates cell chemo-sensitivity by targeting FOXO3 in prostatic cancer. Gene 2018, 658, 152–158.
- Gao, Q.; Zheng, J. microRNA-323 upregulation promotes prostate cancer growth and docetaxel resistance by repressing p73. Biomed. Pharmacother. 2018, 97, 528–534.
- Zhou, W.; Huang, S.; Jiang, Q.; Yuan, T. Suppression of miR-4735-3p in androgen receptor-expressing prostate cancer cells increases cell death during chemotherapy. Am. J. Transl. Res. 2017, 9, 3714–3722.
- Wang, L.; Cho, K.B.; Li, Y.; Tao, G.; Xie, Z.; Guo, B. Long Noncoding RNA (lncRNA)-Mediated Competing Endogenous RNA Networks Provide Novel Potential Biomarkers and Therapeutic Targets for Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 5758.
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigo, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548.
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447.
- Xia, K.G.; Wang, C.M.; Shen, D.Y.; Song, X.Y.; Mu, X.Y.; Zhou, J.W.; Zhu, A.Y.; Xuan, Q.; Tao, T. LncRNA NEAT1-associated aerobic glycolysis blunts tumor immunosurveillance by T cells in prostate cancer. Neoplasma 2022, 69, 594–602.
- Zhao, W.; Zhu, X.; Jin, Q.; Lin, B.; Ji, R. The lncRNA NEAT1/miRNA-766-5p/E2F3 Regulatory Axis Promotes Prostate Cancer Progression. J. Oncol. 2022, 2022, 1866972.
- Jiang, X.; Guo, S.; Zhang, Y.; Zhao, Y.; Li, X.; Jia, Y.; Xu, Y.; Ma, B. LncRNA NEAT1 promotes docetaxel resistance in prostate cancer by regulating ACSL4 via sponging miR-34a-5p and miR-204-5p. Cell Signal. 2020, 65, 109422.
- Wang, X.; Yang, B.; Ma, B. The UCA1/miR-204/Sirt1 axis modulates docetaxel sensitivity of prostate cancer cells. Cancer Chemother. Pharmacol. 2016, 78, 1025–1031.
- Jiang, X.; Guo, S.; Xu, M.; Ma, B.; Liu, R.; Xu, Y.; Zhang, Y. TFAP2C-Mediated lncRNA PCAT1 Inhibits Ferroptosis in Docetaxel-Resistant Prostate Cancer Through c-Myc/miR-25-3p/SLC7A11 Signaling. Front. Oncol. 2022, 12, 862015.
- Ma, Y.; Fan, B.; Ren, Z.; Liu, B.; Wang, Y. Long noncoding RNA DANCR contributes to docetaxel resistance in prostate cancer through targeting the miR-34a-5p/JAG1 pathway. Onco Targets Ther. 2019, 12, 5485–5497.
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018, 11, 63.
- Ren, S.; Liu, Y.; Xu, W.; Sun, Y.; Lu, J.; Wang, F.; Wei, M.; Shen, J.; Hou, J.; Gao, X.; et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J. Urol. 2013, 190, 2278–2287.
- Xue, D.; Lu, H.; Xu, H.Y.; Zhou, C.X.; He, X.Z. Long noncoding RNA MALAT1 enhances the docetaxel resistance of prostate cancer cells via miR-145-5p-mediated regulation of AKAP12. J. Cell Mol. Med. 2018, 22, 3223–3237.
- Liu, X.; Tong, Y.; Xia, D.; Peng, E.; Yang, X.; Liu, H.; Ye, T.; Wang, X.; He, Y.; Ye, Z.; et al. Circular RNAs in prostate cancer: Biogenesis, biological functions, and clinical significance. Mol. Ther. Nucleic Acids 2021, 26, 1130–1147.
- Abdelmohsen, K.; Panda, A.C.; Munk, R.; Grammatikakis, I.; Dudekula, D.B.; De, S.; Kim, J.; Noh, J.H.; Kim, K.M.; Martindale, J.L.; et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017, 14, 361–369.
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e27.
- Nisar, S.; Bhat, A.A.; Singh, M.; Karedath, T.; Rizwan, A.; Hashem, S.; Bagga, P.; Reddy, R.; Jamal, F.; Uddin, S.; et al. Insights Into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies. Front. Cell Dev. Biol. 2021, 9, 617281.
- Zhao, X.; Cai, Y.; Xu, J. Circular RNAs: Biogenesis, Mechanism, and Function in Human Cancers. Int. J. Mol. Sci. 2019, 20, 3926.
- Xin, R.; Gao, Y.; Gao, Y.; Wang, R.; Kadash-Edmondson, K.E.; Liu, B.; Wang, Y.; Lin, L.; Xing, Y. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun. 2021, 12, 266.
- Wu, W.; Ji, P.; Zhao, F. CircAtlas: An integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020, 21, 101.
- Gao, Y.; Liu, J.; Huan, J.; Che, F. Downregulation of circular RNA hsa_circ_0000735 boosts prostate cancer sensitivity to docetaxel via sponging miR-7. Cancer Cell Int. 2020, 20, 334.
- Shen, Z.; Zhou, L.; Zhang, C.; Xu, J. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020, 468, 88–101.
More
Information
Subjects:
Oncology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
617
Revisions:
2 times
(View History)
Update Date:
20 Dec 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




