
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Klaus Ebnet | -- | 1445 | 2023-12-11 10:13:21 | | | |
| 2 | Wendy Huang | Meta information modification | 1445 | 2023-12-11 13:48:39 | | |
Video Upload Options
Tight junctions (TJ) are cell–cell adhesive structures that define the permeability of barrier-forming epithelia and endothelia. In contrast to this seemingly static function, TJs display a surprisingly high molecular complexity and unexpected dynamic regulation, which allows the TJs to maintain a barrier in the presence of physiological forces and in response to perturbations. Cell–cell adhesion receptors play key roles during the dynamic regulation of TJs. They connect individual cells within cellular sheets and link sites of cell–cell contacts to the underlying actin cytoskeleton.
1. Introduction
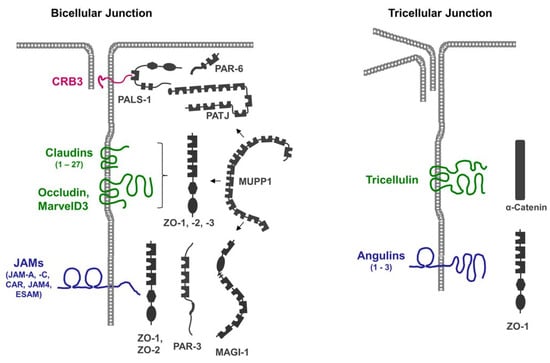
2. CRB3
3. Claudins
4. TAMPs
5. IgSF Proteins
| Integral Membrane Protein | Size (AA) | Cytoplasmic Region (AA) | COOH-Terminal Residues (PBM) |
|---|---|---|---|
| CRB3 | 120 | 40 | - EERLI (+) |
| JAM-A | 299 | 40 | - SSFLV (+) |
| JAM-C | 310 | 48 | - SSFVI (+) |
| CAR | 365 | 107 | - DGSIV (+) |
| JAM4 | 407 | 122 | - NTTVV (+) |
| ESAM | 390 | 121 | - AGSLV (+) |
| Angulin-1/LSR | 649 | 369 | - ESLVV (+) |
| Angulin-2/ILDR1 | 546 | 358 | - RSVVI (+) |
| Angulin-3/ILDR2 | 639 | 432 | - MSLVV (+) |
| Claudins (1–26) | 207–305 | 27–66 | - XXXYF (+) |
| Occludin | 522 | 257 | - DRQKT (−) |
| Tricellulin | 558 | 196 | - VQGYS (−) |
| MarvelD3 | 401 | 20 | - EMFEF (+) |
The presence of multiple PDZ domains in many scaffolding proteins localized at the TJs (Figure 1), combined with the promiscuity of PDZ domains in ligand binding, allows for the incorporation of several integral membrane proteins in a single scaffolding protein-organized complex. Vice versa, individual membrane proteins can recruit and assemble distinct protein complexes at the TJ. These biochemical properties may, in part, explain the high complexity and dynamics of protein complexes at the TJs [21].
References
- Buckley, C.E.; St Johnston, D. Apical-basal polarity and the control of epithelial form and function. Nat. Rev. Mol. Cell Biol. 2022, 23, 559–577.
- Zihni, C.; Mills, C.; Matter, K.; Balda, M.S. Tight junctions: From simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 2016, 17, 564–580.
- Hegazy, M.; Perl, A.L.; Svoboda, S.A.; Green, K.J. Desmosomal Cadherins in Health and Disease. Annu. Rev. Pathol. 2022, 17, 47–72.
- Ladoux, B.; Mege, R.M. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 2017, 18, 743–757.
- Fernandez-Gonzalez, R.; Peifer, M. Powering morphogenesis: Multiscale challenges at the interface of cell adhesion and the cytoskeleton. Mol. Biol. Cell 2022, 33, pe4.
- Knauf, F.; Brewer, J.R.; Flavell, R.A. Immunity, microbiota and kidney disease. Nat. Rev. Nephrol. 2019, 15, 263–274.
- Lambert, A.W.; Weinberg, R.A. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer 2021, 21, 325–338.
- Martin-Belmonte, F.; Perez-Moreno, M. Epithelial cell polarity, stem cells and cancer. Nat. Rev. Cancer 2011, 12, 23–38.
- Miller, P.W.; Clarke, D.N.; Weis, W.I.; Lowe, C.J.; Nelson, W.J. The evolutionary origin of epithelial cell-cell adhesion mechanisms. Curr. Top. Membr. 2013, 72, 267–311.
- Murray, P.S.; Zaidel-Bar, R. Pre-metazoan origins and evolution of the cadherin adhesome. Biol. Open 2014, 3, 1183–1195.
- Abedin, M.; King, N. Diverse evolutionary paths to cell adhesion. Trends Cell Biol. 2010, 20, 734–742.
- Green, K.J.; Roth-Carter, Q.; Niessen, C.M.; Nichols, S.A. Tracing the Evolutionary Origin of Desmosomes. Curr. Biol. 2020, 30, R535–R543.
- Rice, C.; De, O.; Alhadyian, H.; Hall, S.; Ward, R.E. Expanding the Junction: New Insights into Non-Occluding Roles for Septate Junction Proteins during Development. J. Dev. Biol. 2021, 9, 11.
- Takeichi, M. Dynamic contacts: Rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 2014, 15, 397–410.
- Vasileva, E.; Citi, S. The role of microtubules in the regulation of epithelial junctions. Tissue Barriers 2018, 6, 1539596.
- Muller, L.; Hatzfeld, M.; Keil, R. Desmosomes as Signaling Hubs in the Regulation of Cell Behavior. Front. Cell Dev. Biol. 2021, 9, 745670.
- Zihni, C.; Balda, M.S.; Matter, K. Signalling at tight junctions during epithelial differentiation and microbial pathogenesis. J. Cell Sci. 2014, 127, 3401–3413.
- Mendonsa, A.M.; Na, T.Y.; Gumbiner, B.M. E-cadherin in contact inhibition and cancer. Oncogene 2018, 37, 4769–4780.
- Broussard, J.A.; Jaiganesh, A.; Zarkoob, H.; Conway, D.E.; Dunn, A.R.; Espinosa, H.D.; Janmey, P.A.; Green, K.J. Scaling up single-cell mechanics to multicellular tissues—The role of the intermediate filament-desmosome network. J. Cell Sci. 2020, 133, jcs228031.
- Margolis, B. The Crumbs3 Polarity Protein. Cold Spring Harb. Perspect. Biol. 2018, 10, a027961.
- Tan, B.; Yatim, S.; Peng, S.; Gunaratne, J.; Hunziker, W.; Ludwig, A. The Mammalian Crumbs Complex Defines a Distinct Polarity Domain Apical of Epithelial Tight Junctions. Curr. Biol. 2020, 30, 2791–2804.e6.
- Fletcher, G.C.; Lucas, E.P.; Brain, R.; Tournier, A.; Thompson, B.J. Positive feedback and mutual antagonism combine to polarize Crumbs in the Drosophila follicle cell epithelium. Curr. Biol. 2012, 22, 1116–1122.
- Furuse, M.; Tsukita, S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006, 16, 181–188.
- Gunzel, D.; Yu, A.S. Claudins and the modulation of tight junction permeability. Physiol. Rev. 2013, 93, 525–569.
- Kubota, K.; Furuse, M.; Sasaki, H.; Sonoda, N.; Fujita, K.; Nagafuchi, A.; Tsukita, S. Ca(2+)-independent cell-adhesion activity of claudins, a family of integral membrane proteins localized at tight junctions. Curr. Biol. 1999, 9, 1035–1038.
- Furuse, M.; Sasaki, H.; Fujimoto, K.; Tsukita, S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 1998, 143, 391–401.
- Furuse, M.; Sasaki, H.; Tsukita, S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J. Cell Biol. 1999, 147, 891–903.
- Tsukita, S.; Tanaka, H.; Tamura, A. The Claudins: From Tight Junctions to Biological Systems. Trends Biochem. Sci. 2019, 44, 141–152.
- Raleigh, D.R.; Marchiando, A.M.; Zhang, Y.; Shen, L.; Sasaki, H.; Wang, Y.; Long, M.; Turner, J.R. Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol. Biol. Cell 2010, 21, 1200–1213.
- Sanchez-Pulido, L.; Martin-Belmonte, F.; Valencia, A.; Alonso, M.A. MARVEL: A conserved domain involved in membrane apposition events. Trends Biochem. Sci. 2002, 27, 599–601.
- Ikenouchi, J.; Furuse, M.; Furuse, K.; Sasaki, H.; Tsukita, S.; Tsukita, S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J. Cell Biol. 2005, 171, 939–945.
- Cording, J.; Berg, J.; Kading, N.; Bellmann, C.; Tscheik, C.; Westphal, J.K.; Milatz, S.; Gunzel, D.; Wolburg, H.; Piontek, J.; et al. In tight junctions, claudins regulate the interactions between occludin, tricellulin and marvelD3, which, inversely, modulate claudin oligomerization. J. Cell Sci. 2013, 126, 554–564.
- Van Itallie, C.M.; Anderson, J.M. Occludin confers adhesiveness when expressed in fibroblasts. J. Cell Sci. 1997, 110 Pt 9, 1113–1121.
- Saitou, M.; Furuse, M.; Sasaki, H.; Schulzke, J.D.; Fromm, M.; Takano, H.; Noda, T.; Tsukita, S. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol. Biol. Cell 2000, 11, 4131–4142.
- Steed, E.; Rodrigues, N.T.; Balda, M.S.; Matter, K. Identification of MarvelD3 as a tight junction-associated transmembrane protein of the occludin family. BMC Cell Biol. 2009, 10, 95.
- Sugawara, T.; Furuse, K.; Otani, T.; Wakayama, T.; Furuse, M. Angulin-1 seals tricellular contacts independently of tricellulin and claudins. J. Cell Biol. 2021, 220, e202005062.
- Cho, Y.; Haraguchi, D.; Shigetomi, K.; Matsuzawa, K.; Uchida, S.; Ikenouchi, J. Tricellulin secures the epithelial barrier at tricellular junctions by interacting with actomyosin. J. Cell Biol. 2022, 221, e202009037.
- Ebnet, K. Junctional Adhesion Molecules (JAMs): Cell Adhesion Receptors With Pleiotropic Functions in Cell Physiology and Development. Physiol. Rev. 2017, 97, 1529–1554.
- Cohen, C.J.; Shieh, J.T.; Pickles, R.J.; Okegawa, T.; Hsieh, J.T.; Bergelson, J.M. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. USA 2001, 98, 15191–15196.
- Hirabayashi, S.; Tajima, M.; Yao, I.; Nishimura, W.; Mori, H.; Hata, Y. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol. Cell. Biol. 2003, 23, 4267–4282.
- Raschperger, E.; Engstrom, U.; Pettersson, R.F.; Fuxe, J. CLMP, a novel member of the CTX family and a new component of epithelial tight junctions. J. Biol. Chem. 2004, 279, 796–804.
- Nasdala, I.; Wolburg-Buchholz, K.; Wolburg, H.; Kuhn, A.; Ebnet, K.; Brachtendorf, G.; Samulowitz, U.; Kuster, B.; Engelhardt, B.; Vestweber, D.; et al. A transmembrane tight junction protein selectively expressed on endothelial cells and platelets. J. Biol. Chem. 2002, 277, 16294–16303.
- Hirata, K.; Ishida, T.; Penta, K.; Rezaee, M.; Yang, E.; Wohlgemuth, J.; Quertermous, T. Cloning of an immunoglobulin family adhesion molecule selectively expressed by endothelial cells. J. Biol. Chem. 2001, 276, 16223–16231.
- Furuse, M.; Izumi, Y.; Oda, Y.; Higashi, T.; Iwamoto, N. Molecular organization of tricellular tight junctions. Tissue Barriers 2014, 2, e28960.
- Higashi, T.; Tokuda, S.; Kitajiri, S.; Masuda, S.; Nakamura, H.; Oda, Y.; Furuse, M. Analysis of the ‘angulin’ proteins LSR, ILDR1 and ILDR2--tricellulin recruitment, epithelial barrier function and implication in deafness pathogenesis. J. Cell Sci. 2013, 126, 966–977.
- Masuda, S.; Oda, Y.; Sasaki, H.; Ikenouchi, J.; Higashi, T.; Akashi, M.; Nishi, E.; Furuse, M. LSR defines cell corners for tricellular tight junction formation in epithelial cells. J. Cell Sci. 2011, 124, 548–555.
- Tonikian, R.; Zhang, Y.; Sazinsky, S.L.; Currell, B.; Yeh, J.H.; Reva, B.; Held, H.A.; Appleton, B.A.; Evangelista, M.; Wu, Y.; et al. A specificity map for the PDZ domain family. PLoS Biol. 2008, 6, e239.




