Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sarfaraz K. Niazi | -- | 3391 | 2023-12-06 07:49:21 | | | |
| 2 | Sirius Huang | Meta information modification | 3391 | 2023-12-08 02:08:06 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Niazi, S.K.; Magoola, M. Escherichia coli-Based Therapeutic Protein Expression. Encyclopedia. Available online: https://encyclopedia.pub/entry/52412 (accessed on 07 February 2026).
Niazi SK, Magoola M. Escherichia coli-Based Therapeutic Protein Expression. Encyclopedia. Available at: https://encyclopedia.pub/entry/52412. Accessed February 07, 2026.
Niazi, Sarfaraz K., Matthias Magoola. "Escherichia coli-Based Therapeutic Protein Expression" Encyclopedia, https://encyclopedia.pub/entry/52412 (accessed February 07, 2026).
Niazi, S.K., & Magoola, M. (2023, December 06). Escherichia coli-Based Therapeutic Protein Expression. In Encyclopedia. https://encyclopedia.pub/entry/52412
Niazi, Sarfaraz K. and Matthias Magoola. "Escherichia coli-Based Therapeutic Protein Expression." Encyclopedia. Web. 06 December, 2023.
Copy Citation
Therapeutic proteins treat many acute and chronic diseases that were, until recently, considered untreatable. However, their high development cost keeps them out of reach of most patients around the world. One possible way to make manufacturing cheaper is to use newer technologies, such as Escherichia coli to make larger molecules, like full-length antibodies, that are normally only made in Chinese Hamster Ovary (CHO) cells, switch to continuous manufacturing, and change the process to cell-free synthesis. The advantages of using E. coli include a shorter production cycle, little risk of viral contamination, cell host stability, and a highly reproducible post-translational modification.
biosimilars
Escherichia coli
recombinant proteins
monoclonal antibody
bispecific antibody
inclusion body
continuous manufacturing (CM)
cell-free protein synthesis (CFPS)
1. Introduction
Therapeutic proteins represent a diverse class of drugs first made accessible as recombinant DNA (rDNA) insulin in 1982 [1]. There are now 266 such approved proteins [2], comprising a wide range of products with unique mechanisms of action and size, ranging from peptides to monoclonal antibodies (Table 1).
Table 1. FDA-approved therapeutic proteins as of July 2023. (https://drugs.ncats.io/, accessed on 10 July 2023).
| Listed Protein Class | Number |
|---|---|
| Monoclonal Antibody | 94 |
| Hormone | 10 |
| Enzyme | 8 |
| Monoclonal Antibody Conjugate | 8 |
| Cytokine | 4 |
| Bispecific Antibody | 3 |
| Coagulation Factor | 3 |
| Growth Factor | 3 |
| Peptide | 3 |
| Carrier Protein | 1 |
| Enzyme Inhibitor | 1 |
| Fab | 1 |
| Fusion Proteins | 1 |
| Single-Domain Antibody | 1 |
| Toxin | 1 |
It is noteworthy that the European Medicines Agency (EMA) lists peptides as proteins, unlike the US Food and Drug Administration (FDA) [3]. Hormones, cytokines, enzymes, antibody fragments, and shorter-than-full-length antibodies that were made by E. coli were used in medicine long before Chinese Hamster Ovary (CHO) cells were used (Table 1). Some proteins still extracted from tissues can also be manufactured using E. coli. Examples include Alpha-1 antitrypsin, Antithrombin III, Botulinum toxin, C1 inhibitor, Fibrinogen, Heparin, Hirudin, Snake venom proteins, Streptokinase, Thrombin, and Urokinase. Figure 1 shows that most proteins expressed in E. coli are of lower molecular weight since most of the more complex and higher molecular weight monoclonal antibodies (mAbs) are expressed in CHO cells.
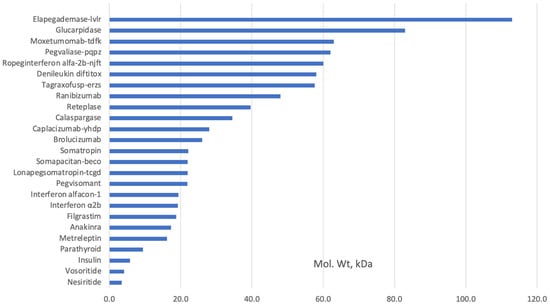
Figure 1. Examples of FDA-approved recombinant proteins produced in E. coli (www.fda.hhs.gov, accessed on 10 July 2023).
The FDA-approved therapeutic proteins expressed in E. coli are primarily of molecular weight of less than 32 kDa; it is difficult to produce proteins higher than 100 kDa in E. coli as it places an excessive cell host load that prevents correct protein folding while maintaining adequate expression levels [4].
The value of E. coli will become more apparent when we start using it to express larger molecules, such as the mAbs. The first study on the expression of full-length (FL) immunoglobulins (FL-IgGs) in E. coli was published in 2002. Later, in 2020, a modular system-based synthetic biology technique was applied to knock down gene expression using short regulatory ribonucleic acids (RNAs) with cetuximab as a target FL-IgG for enhancing expression, reaching up to 200 mg/L [5].
Combining E. coli with emerging technologies like bioinformatics, novel methods for genetic manipulation to force E. coli to secrete heterologous proteins, and managing post-translational modification offer many new opportunities, including E. coli-based continuous manufacturing and cell-free synthesis, that can significantly reduce the cost of development and manufacturing as well as enhance product safety.
2. Background
To make recombinant DNA products, DNA from different species are joined together and then put into a host cell, usually a bacterium or mammalian cell, to make the protein that is wanted. The pioneering development of this molecular chimera dates back to 1972 [6][7] when researchers affiliated with the University of California, San Francisco, and Stanford University accomplished this technique. The United States patent for the invention was granted to Stanley Cohen from Stanford University and Herbert Boyer from the University of California, San Francisco (UCSF) in 1980. In 1976, Boyer played a crucial role in establishing Genentech, Inc. These patents have been licensed to more than 500 licensees and yielded royalties exceeding USD 250 million for Stanford and UCSF [8].
Since the FDA approved the first recombinant protein for therapeutic purposes in 1982, E. coli has remained a prominent organism for producing recombinant proteins despite the availability of many newer expression systems. Using microbial expression systems, especially E. coli, to make even heterologous recombinant proteins is still an easier and cheaper way to do it than using mammalian cell culture or other systems. E. coli presents several notable benefits in genetic manipulation, growth conditions, high product yields, product purity, absence of viral contamination, and many more.
3. Bioinformatics Applications
A methodical process (Figure 2) is needed to decide if E. coli is a good system and if using it will help reach the goal of lower-cost manufacturing. The first step is to look at a number of factors, including possible splice variants, signal sequences, transmembrane helices, and post-translational modifications seen in the native protein. Protein databases, such as UniProt [9], are valuable initial bioinformatics resources, although they remain suggestive and not definitive. For example, knowing what can be produced in E. coli and what cannot is critical knowledge; cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) is an obligate dimer and requires N-glycosylation of Asn78 and Asn110 for dimerization [10], and this PTM cannot be made in E. coli. This will need a synthetic biology method since the only eukaryotic-like PTMs that E. coli can produce is disulfide bond formation in the periplasm [11].
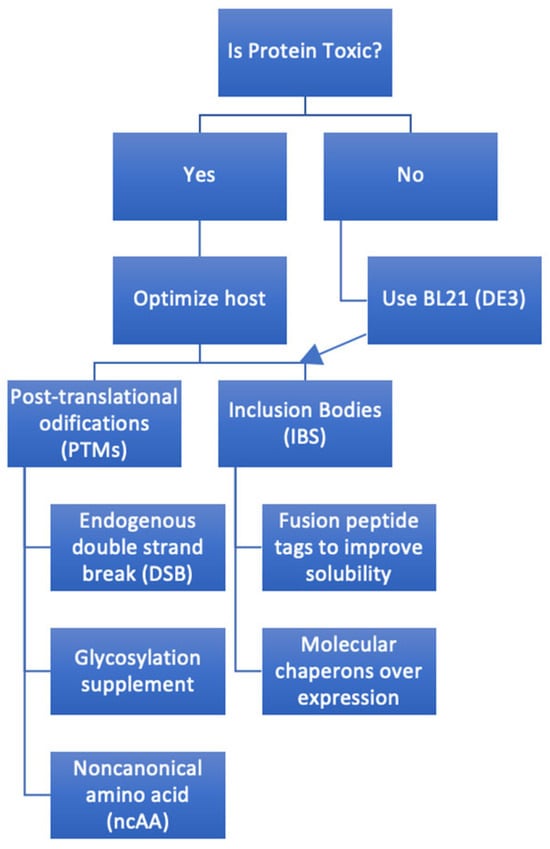
Figure 2. A systematic process of creating a plan to express a full-length antibody in E. coli. Arrow indicates optimized host follow-on.
Bioinformatics methodologies, such as the software JPRED 4 [12], facilitate the investigation of domain boundaries and the prediction of regions of intrinsically disordered proteins (IDPs). Failure to express a construct that is insufficient in length and lacks a crucial component within a certain domain, such as a β-strand, should be anticipated. On the other hand, trying to express a construct that is too long and includes flexible parts that can be broken down by proteases will probably result in either heterogeneity or the loss of a purification tag. Proteins with a lot of intrinsically disordered regions (IDRs) can be hard to make because they break down easily. Proteins characterized by a significant proportion of intrinsically disordered regions (IDRs) often provide challenges in their production due to their inherent susceptibility to degradation. However, these may become structured upon interaction with other molecules, forming complexes such as acetyltransferase (ACTR) and nuclear coactivator binding domain (NCBD) [13] that can help as partners, making a stable and soluble protein.
Expanding the scope of the use of E. coli systems is now based on several well-defined prospects:
-
Exploiting the use of bioinformatics tools to determine the biophysical characteristics of the protein [14]. It is a complex process that involves various computational methods. These methods utilize algorithms and statistical models to analyze the protein’s primary sequence, infer its three-dimensional structure, and predict its interactions and functions.
- ○
-
Sequence analysis involves comparing the amino acid sequence of a protein with known sequences in databases to identify conserved domains, motifs, or families [15];
- ○
-
Structure prediction includes methods like homology modeling, ab initio modeling, and threading to predict a protein’s three-dimensional (3D) structure based on its sequence [16];
- ○
-
Functional prediction identifies the biological role of a protein by assessing its structural and sequential features, often in conjunction with known protein-protein interactions and pathway analyses [17];
- ○
-
Molecular dynamics simulations and related techniques are used to study the movement and interactions of proteins, providing insight into their behavior in the cellular environment [18];
- ○
-
Specific bioinformatics tools are designed to predict sites in proteins likely to undergo post-translational modifications (PTMs) such as phosphorylation or glycosylation [19];
- ○
-
Predicting how proteins interact with other proteins or ligands can be achieved through docking simulations and other modeling techniques [20].
-
Accurate delineation [21]:
- ○
-
Identifying the boundaries of protein domains is essential for understanding the function and evolution of proteins [22];
- ○
-
Signal sequences are crucial for the targeting of proteins to specific cellular locations. Identifying these sequences helps in understanding the transportation and localization of proteins [23];
- ○
-
Transmembrane regions anchor proteins in membranes, playing essential roles in cellular communication, signaling, and transport. Accurate prediction of these regions aids in understanding membrane protein structure and function [24];
- ○
-
Identifying obligate oligomeric complexes is essential for understanding protein-protein interactions and the assembly of multi-protein complexes [25];
- ○
-
Identification of PTMs is vital for understanding protein regulation and signaling [26].
Optimization of genetic and translation variables encompasses various elements, including codon use, the characteristics and placement of the ribosome binding site, and disparities in translation rates between prokaryotes and eukaryotes [27].
4. Gene Cloning and Design
Gene cloning typically involves selecting a purification method, such as affinity chromatography, which utilizes the inherent characteristics of the protein. This can be achieved through immobilized ligand or substrate mimic chromatography, using compounds like Cibacron Blue F3GA [28] or cyclic peptide-based ligands [29]. Alternatively, a purification tag, such as a maltose-binding protein (MBP)-tag, glutathione-S-transferase (GST)-tag, or commonly a hexahistidine tag (his-tag), can be added to facilitate purification. Immobilized metal affinity chromatography (IMAC) [30] is commonly employed. If a protein with similar characteristics is present, its attributes can be utilized to assess the feasibility of adding a tag to the N- and C-terminus. Alternatively, one might utilize structure prediction software such as Phyre 2 [31]. Although N-terminal histidine tags are highly valuable and extensively employed, they can introduce heterogeneity in the final product due to varying (phospho)gluconylation occurring at the N-terminus [32].
After the design of the protein construct, gene design starts to yield maximum expression that depends much on cellular homeostasis, or keeping a delicate balance within the cell. When a high-copy number plasmid is employed with a robust promoter, it consistently leads to a reduced protein yield [33]. This is attributed to the excessive allocation of cellular resources towards synthesizing plasmid DNA and mRNA. Consequently, the abundance of mRNA exceeds the capacity of the translation machinery, resulting in suboptimal protein production. Toxic effects of overexpressed recombinant proteins on E. coli cells can be anticipated to avoid these processes [34].
Transcriptome analysis can identify and remove the genes in charge of the cellular stress response. The number of growth-essential genes’ down-regulated expression is reduced when cell surface receptor (CSR) is blocked [35].
An increasingly popular method to avoid losing plasmids during long fermentation processes is to add genes to the bacterial chromosome. However, despite their drawbacks, plasmids can be employed in their original form because they are more expeditious and cost-effective. The selection of plasmids for protein production depends on their copy quantity, which depends on the plasmid's origin of replication, promoter, and selection marker. To get the most out of the cell resources used for protein production, you need to find the right balance between the number of copies of the plasmid and the strength of the promoter, taking into account the conditions of the media.
The field of synthetic biology has witnessed notable progress in developing growth-decoupled recombinant protein production. This has been achieved using the co-expression of Gp2, a peptide generated from a bacteriophage that acts as an inhibitor of RNA polymerase in Escherichia coli. This methodology facilitated the regulation of metabolic resources, ensuring their exclusive allocation towards synthesizing the intended protein.
In addition to the plasmid, the origin of the gene is a crucial factor. In the past, the gene was taken directly from the living thing itself, usually by using a cDNA library made from messenger RNA (mRNA) through reverse transcription polymerase chain reaction (RT-PCR) to avoid including introns. Although the process can exhibit rapidity, cost-effectiveness, and efficiency, it can also lead to challenges associated with disparities in translation initiation and codon utilization between prokaryotic and eukaryotic organisms.
Due to a significant decrease in pricing, the cost of synthesizing a gene artificially has become lower than the combined expenses of labor and materials involved in cloning a gene from a complementary DNA (cDNA) library. Synthetic genes can also alleviate the potentially harmful consequences of another dissimilarity in protein translation rates between eukaryotes and prokaryotes [36]. In prokaryotic organisms like Escherichia coli (E. coli), a coupling exists between the transcription and translation rates [37]. Specifically, transcription occurs at a rate of 50 nucleotides, whereas translation occurs at 16 amino acids.
4.1. Ribosomes
In 1987 [38], a modified ribosome system was developed to facilitate the production of the proteins in E. coli through modifications made to the Shine–Dalgarno (SD) sequence of the mRNA and the corresponding anti-SD sequence of the 16S ribosomal RNA (rRNA). Other alternative ribosome systems can be utilized, including the orthogonal riboswitch system [39], the RiboTite system, and the Ribo-T system [40][41]. The riboswitch system facilitates the adjustable co-expression of several genes in a dose-dependent manner in response to tiny synthetic chemicals. On the other hand, the RiboTite system, an extension of the riboswitch technology, has demonstrated the ability to synchronize protein translation rates with protein release. The Ribo-T system utilizes a modified hybrid rRNA that combines small and large subunit rRNA sequences. This modified rRNA is connected into a single translating unit using short RNA linkers that form covalent bonds between the subunits. The functionality of the orthogonal ribosome-mRNA system has been demonstrated to sustain bacterial growth in the absence of wild-type ribosomes. Furthermore, a recent study has documented the development of an enhanced tethered version of this system [42].
-
The characteristics and location of the ribosome binding site (RBS) and the disparities in translation rates observed in prokaryotic and eukaryotic organisms [43]. The ribosome binding site (RBS) plays a crucial role in translation initiation. The sequence and position of a gene relative to the initiation codon can influence translation efficiency. Customizing the RBS to the host organism might enhance the efficiency of translating the desired protein [44];
-
Correct use of the strain and media to optimize production, though with many limitations [45]. The optimization of production in E. coli strains through proper selection of the strain and media is a common strategy in biotechnology but comes with certain limitations.
-
Optimization in E. coli can vary widely depending on the protein or other manufactured product. Selecting the right strain of E. coli, determining the optimal temperature, and choosing the appropriate culture media are crucial considerations for recombinant protein expression.
The presence of secondary structural components in mRNA might obstruct ribosome binding, resulting in hindered translation and various limits in the translational process [46]. Eukaryotic ribosomes exhibit a binding affinity towards the cap located at the 5′ terminus of the mRNA molecule. Subsequently, they traverse along the mRNA until they commence translation at the initial AUG codon, preceded by a Kozak sequence. In contrast, prokaryotic ribosomes engage with a specific region on the mRNA called the Shine–Dalgarno sequence or ribosome binding site. The ribosome binding sites (RBS) typically consist of 5–13 base pairs [47] upstream of the beginning AUG codon, with an ideal spacing of 5–6 base pairs [48]. These RBS sequences complement the 3′ end of the 16S ribosomal RNA. The nucleotide sequence AGGAGGU [49] is seen in Escherichia coli. When eukaryotic proteins are made in Escherichia coli (E. coli), having a separate ribosome binding site (RBS) causes two different things to happen. Before beginning the AUG codon, a ribosome binding site (RBS) must be present. This phenomenon may be observed within the plasmid region external to the multi-cloning site. However, it is imperative to exercise caution to ensure that the distance is appropriate and that the translation process does not inadvertently introduce more AUG trinucleotides.
Furthermore, it is essential that this specific nucleotide sequence does not occur inside the gene of interest. At an internal ribosome binding site (RBS), two things can happen: if there is an AUG codon close enough to it, it can either cause translation to stop because a ribosome binds to it and stops translation; or it can cause the production of a second protein. Therefore, special consideration is given to the choice of codons for Gly-Gly pairs (excluding GGA-GGU), Arg-Arg pairs (excluding AGG-AGG), and sequences surrounding Glu (GAG), including Glu-Glu pairs (GAG-GAG). Escherichia coli (E. coli) exhibits infrequent utilization of AGG and GGA codons. Because of this, it is very important to be careful when optimizing codons to avoid internal ribosome binding sites (RBS) that are linked to sequences around glutamic acid (Q/K/E-E or E-V).
4.2. Promoter
Some important functional parts close to PT7 are the −35/−10 region, the translation initiation region (TIR), the operator sequence, and the TpET plasmid's replicon. There are many functional areas close to PT7, which is the core region of the pET plasmid, that control the level of expression before induction and the right transcription rate after induction.
By maximizing transcription or translation levels, the T7 RNAP objective is attained. The lacUV5 promoter (PlacUV5), a strongly inducible promoter that is activated by the amino acid isopropyl-beta-d-thiogalactopyranoside (IPTG), controls this process [50], and the PlacUV5 is independent of recombinant product, which makes it leakier than Plac [51]. Three inducible promoters—ParaBAD [52], PrhaBAD, and Ptet—are appropriate for toxin–protein fermentation that lasts a long time. PrhaBAD and Ptet, however, more strictly control T7 RNAP transcription, giving additional expression possibilities for various recombinant products—especially dangerous proteins [53]. When the lac repressor gene (lacI) is altered, leaky expression is decreased by improving the ability to inhibit proteins [54].
-
To create the promoter variation lac1G, the promoter lacUV5 and lac were joined again. (G was substituted for A at position +1) [55];
-
By having a mutant form of the Lac repressor protein (LacI), specifically the V192F variant, the expression of T7 RNA polymerase (RNAP) is effectively controlled to stop leakage. This mutant variant cannot bind to isopropyl β-D-1-thiogalactopyranoside (IPTG), hence preventing its activation. Consequently, the mutant LacI dynamically governs the levels of transcripts produced by T7 RNAP [56];
-
Building a T7 RNAP RBS library quickly involves using the base editor and CRISPR/Cas9 to screen potential expression hosts [57];
-
Because of a specific amino acid substitution (A102D), T7 RNA polymerase was less able to bind to the PT7 promoter. This changed the rate at which RNA was made. The T7 RNA polymerase (T7 RNAP) was fragmented into two segments and co-expressed with a light-responsive dimerization domain, exhibiting functional behavior upon exposure to blue light [58].
4.3. Codons
The expression level of the ColE1 plasmid replication-associated gene can be regulated by utilizing CRISPRi and the inducible promoter Ptet [59].
The distribution of codon usage is not uniform throughout the available codons, and there is significant variance in the degree of codon usage bias observed among different organisms. Using codons exhibits substantial variation across other microorganisms and is associated with corresponding transfer RNA (tRNA) quantities [60].
mRNA, which contains multiple rare codons, can exhibit translation stalling and degradation [61]. Bioinformatic approaches can examine codon usage issues, e.g., Graphical Codon Usage Analyzer [62]. One method to prevent this problem is to overexpress the rare tRNAs [63], such as from pLysSRARE [64][65]. The usual approach is using synthetic genes that can be codon optimized for the expression host while avoiding internal RBS, internal restriction sites, and factors that influence mRNA structure and stability [66][67].
4.4. Protein Folding
Translation rates in eukaryotes are comparatively slower, typically occurring at approximately three amino acids per second. The process of protein folding has co-evolved with translation rates, resulting in a situation where the translation rate [68] of a eukaryotic protein expressed in E. coli may exceed the folding rate. This poses a challenge, particularly for multi-domain proteins. However, this challenge can be addressed through various strategies, such as adjusting the translation rate, harmonizing codon usage [69], or intentionally inducing ribosome stalling by incorporating rarer codons at domain boundaries.
When the host cell cannot handle the rate or volume of recombinant products being expressed, many proteins will misfold and cluster, eventually creating IBs and obstructing the expression. The primary reasons for the synthesis of IBs are limited post-translational modifications (PTMs) capacity and folding efficiency, which are of the utmost importance for increasing the functional activity of recombinant products [70].
To make sure that antibodies with disulfide linkages fold and work correctly, the individual antibody chains need to be exposed to the oxidizing conditions in the periplasm of bacteria. In addition, it should be noted that the periplasmic space serves as a habitat for specific proteins known as chaperonins and disulfide isomerases, which play a crucial role in correctly folding newly synthesized proteins [71]. A leader sequence (PelB, OmpA, PhoA) drives the antibody to the oxidizing periplasm for periplasmic expression [72]. After expression, osmotic shock extracts the antibody from the periplasmic region. Yields obtained from shaking flask cultures have been documented to range from 0.1 mg/L to 100 mg/L, while using fermenters has demonstrated the potential to achieve yields as high as 2 g/L [73]. Utilizing specific E. coli strains that offer an oxidizing environment in the cytoplasm is an additional choice; typically, it comprises mutations of the enzymes, glutathione oxidoreductases, and thioredoxin reductases [74].
Choosing the right molecular chaperones, such as GroES/GroEL, DnaK-DnaJ-GrpE, and co-expression, for overexpression to increase folding efficiency [75].
References
- Landgraf, W.; Sandow, J. Recombinant Human Insulins–Clinical Efficacy and Safety in Diabetes Therapy. Eur. Endocrinol. 2016, 12, 12–17.
- Approved Protein Drugs in the US and EU. Available online: https://drugs.ncats.io/ (accessed on 10 July 2023).
- Dimitrov, D.S. Therapeutic proteins. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 899, pp. 1–26.
- Zhang, Z.X.; Nong, F.T.; Wang, Y.Z.; Yan, C.-X.; Gu, Y.; Song, P.; Sun, X.-M. Strategies for efficient production of recombinant proteins in Escherichia coli: Alleviating the host burden and enhancing protein activity. Microb. Cell Fact. 2022, 21, 191.
- Zhang, J.; Zhao, Y.; Cao, Y.; Yu, Z.; Wang, G.; Li, Y.; Ye, X.; Li, C.; Lin, X.; Song, H. Synthetic sRNA-based engineering of Escherichia coli for enhanced production of full-length immunoglobulin G. Biotechnol. J. 2020, 15, e1900363.
- Jackson, D.A.; Symons, R.H.; Berg, P. Biochemical method for inserting new genetic information into DNA of Simian Virus 40: Circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proc. Natl. Acad. Sci. USA 1972, 69, 2904–2909.
- Cohen, S.N.; Chang, A.C.; Boyer, H.W.; Helling, R.B. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. USA 1973, 70, 3240–3244.
- Feldman, M.P.; Colaianni, A.; Liu, K. Lessons from the Commercialization of the Cohen-Boyer Patents: The Stanford University Licensing Program. Handbook of Best Practices. In Intellectual Property Management in Health and Agricultural Innovation: A Handbook of Best Practices; MIHR: Oxford, UK; PIPRA: Davis, CA, USA, 2007; Available online: https://maryannfeldman.web (accessed on 10 July 2023).
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515.
- Darlington, P.J.; Kirchhof, M.G.; Criado, G.; Sondhi, J.; Madrenas, J. Hierarchical Regulation of CTLA-4 Dimer-Based Lattice Formation and Its Biological Relevance for T Cell Inactivation. J. Immunol. 2005, 175, 996–1004.
- Manta, B.; Boyd, D.; Berkmen, M. Disulfide Bond Formation in the Periplasm of Escherichia coli. EcoSal Plus 2019, 8, 10–1128.
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394.
- Demarest, S.J.; Martinez-Yamout, M.; Chung, J.; Chen, H.; Xu, W.; Dyson, H.J.; Evans, R.M.; Wright, P.E. Mutual synergistic folding in recruitment of cbp/p300 by p160 nuclear receptor coactivators. Nature 2002, 415, 549–553.
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710.
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410.
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 1999, 12, 85–94.
- Radivojac, P.; Clark, W.T.; Oron, T.R.; Schnoes, A.M.; Wittkop, T.; Sokolov, A.; Graim, K.; Funk, C.; Verspoor, K.; Ben-Hur, A.; et al. A large-scale evaluation of computational protein function prediction. Nat. Methods 2013, 10, 221–227.
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652.
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999, 294, 1351–1362.
- Vakser, I.A. Protein docking for low-resolution structures. Protein Eng. 1995, 8, 371–377.
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 5th ed.; Garland Science: New York, NY, USA, 2008; ISBN 978-0-8153-4105-5.
- Sillitoe, I.; Lewis, T.E.; Cuff, A.; Das, S.; Ashford, P.; Dawson, N.L.; Furnham, N.; Laskowski, R.A.; Lee, D.; Lees, J.G.; et al. CATH: Comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 2015, 43, D376–D381.
- von Heijne, G. The signal peptide. J. Membr. Biol. 1990, 115, 195–201.
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580.
- Levy, E.D.; Teichmann, S.A. Structural, evolutionary, and assembly principles of protein oligomerization. Prog. Mol. Biol. Transl. Sci. 2013, 117, 25–51.
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J., Jr. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew. Chem. Int. Ed. 2005, 44, 7342–7372.
- Kudla, G.; Murray, A.W.; Tollervey, D.; Plotkin, J.B. Coding-sequence determinants of gene expression in Escherichia coli. Science 2009, 324, 255–258.
- Subramanian, S.; Ross, P.D. Dye-ligand affinity chromatography: The interaction of cibacron blue f3GA® with proteins and enzyme. Crit. Rev. Biochem. Mol. Biol. 1984, 16, 169–205.
- Kish, W.S.; Roach, M.K.; Sachi, H.; Naik, A.D.; Menegatti, S.; Carbonell, R.G. Purification of human erythropoietin by affinity chromatography using cyclic peptide ligands. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018, 1085, 1–12.
- Young, C.L.; Britton, Z.T.; Robinson, A.S. Recombinant protein expression and purification: A comprehensive review of affinity tags and microbial applications. Biotechnol. J. 2012, 7, 620–634.
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858.
- Geoghegan, K.F.; Dixon, H.B.F.; Rosner, P.J.; Hoth, L.R.; Lanzetti, A.J.; Borzilleri, K.A.; Marr, E.S.; Pezzullo, L.H.; Martin, L.B.; LeMotte, P.K.; et al. Spontaneous α-N-6-phosphogluconoylation of a “His tag” in Escherichia coli: The cause of extra mass of 258 or 178 Da in fusion proteins. Anal. Biochem. 1999, 267, 169–184.
- Wood, W.N.; Smith, K.D.; Ream, J.A.; Lewis, L.K. Enhancing yields of low and single copy number plasmid DNAs from Escherichia coli cells. J. Microbiol. Methods 2017, 133, 46–51.
- Jeong, K.J.; Lee, S.Y. High-level production of human leptin by fed-batch cultivation of recombinant Escherichia coli and its purification. Appl. Environ. Microbiol. 1999, 65, 3027–3032.
- Sharma, A.K.; Shukla, E.; Janoti, D.S.; Mukherjee, K.J.; Shiloach, J. A novel knock out strategy to enhance recombinant protein expression in Escherichia coli. Microb. Cell Fact. 2020, 19, 148.
- Ganoza, M.C.; Kiel, M.C.; Aoki, H. Evolutionary conservation of reactions in translation. Microbiol. Mol. Biol. Rev. 2002, 66, 460–485.
- Irastortza-Olaziregi, M.; Amster-Choder, O. Coupled Transcription-Translation in Prokaryotes: An Old Couple with New Surprises. Front. Microbiol. 2021, 11, 624830.
- Hui, A.; De Boer, H.A. Specialized ribosome system: Preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. USA 1987, 84, 4762–4766.
- Dixon, N.; Robinson, C.J.; Geerlings, T.; Duncan, J.N.; Drummond, S.P.; Micklefield, J. Orthogonal Riboswitches for Tuneable Coexpression in Bacteria. Angew. Chem. Int. Ed. 2012, 51, 3620–3624.
- Orelle, C.; Carlson, E.D.; Szal, T.; Florin, T.; Jewett, M.C.; Mankin, A.S. Protein synthesis by ribosomes with tethered subunits. Nature 2015, 524, 119–124.
- Morra, R.; Shankar, J.; Robinson, C.J.; Halliwell, S.; Butler, L.; Upton, M.; Hay, S.; Micklefield, J.; Dixon, N. Dual transcriptional-Translational cascade permits cellular level tuneable expression control. Nucleic Acids Res. 2016, 44, 21.
- Carlson, E.D.; d’Aquino, A.E.; Kim, D.S.; Fulk, E.M.; Hoang, K.; Szal, T.; Mankin, A.S.; Jewett, M.C. Engineered ribosomes with tethered subunits for expanding biological function. Nat. Commun. 2019, 10, 3920.
- Sharp, P.M.; Li, W.H. The codon Adaptation Index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295.
- Salis, H.M.; Mirsky, E.A.; Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950.
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172.
- Zhang, G.; Darst, S.A. Structure of the Escherichia coli RNA polymerase α subunit amino-terminal domain. Science 1998, 281, 262–266.
- Chen, H.; Bjerknes, M.; Kumar, R.; Jay, E. Determination of the optimal aligned spacing between the shine-dalgarno sequence and the translation initiation codon of Escherichia coli m RNAs. Nucleic Acids Res. 1994, 22, 4953–4957.
- Shepard, H.M.; Yelverton, E.; Goeddel, D.V. Increased Synthesis in E. coli of Fibroblast and Leukocyte Interferons Through Alterations in Ribosome Binding Sites. DNA 1982, 1, 125–131.
- Shine, J.; Dalgarno, L. The 3′ terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 1974, 71, 1342–1346.
- Jeong, H.; Barbe, V.; Lee, C.H.; Vallenet, D.; Yu, D.S.; Choi, S.-H.; Couloux, A.; Lee, S.-W.; Yoon, S.H.; Cattolico, L. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 2009, 394, 644–652.
- Du, F.; Liu, Y.-Q.; Xu, Y.S.; Li, Z.J.; Wang, Y.Z.; Zhang, Z.X.; Sun, X.M. Regulating the T7 RNA polymerase expression in E. coli BL21(DE3) to provide more host options for recombinant protein production. Microb. Cell Fact. 2021, 20, 189.
- Khlebnikov, A.; Risa, Ø.; Skaug, T.; Carrier, T.A.; Keasling, J.D. Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture. J. Bacteriol. 2000, 182, 7029–7034.
- Lutz, R.; Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997, 25, 1203–1210.
- Mueller, K.L.; Simon, J.D.; Elf, J. Design, construction, and implementation of a fully repressible bistable genetic switch in E. coli. Nucleic Acids Res. 2019, 47, 6307–6317.
- Sun, X.M.; Zhang, Z.X.; Wang, L.R.; Wang, J.G.; Liang, Y.; Yang, H.F.; Tao, R.S.; Jiang, Y.; Yang, J.J.; Yang, S. Downregulation of T7 RNA polymerase transcription enhances pET-based recombinant protein production in Escherichia coli BL21(DE3) by suppressing autolysis. Biotechnol. Bioeng. 2021, 118, 153–163.
- Kim, S.K.; Lee, D.-H.; Kim, O.C.; Kim, J.F.; Yoon, S.H. Tunable control of an Escherichia coli expression system for the overproduction of membrane proteins by titrated expression of a mutant lac repressor. ACS Synth. Biol. 2017, 6, 1766–1773.
- Li, Z.J.; Zhang, Z.X.; Xu, Y.; Shi, T.Q.; Ye, C.; Sun, X.M.; Huang, H. CRISPR-Based Construction of a BL21 (DE3)-derived variant strain library to rapidly improve recombinant protein production. ACS Synth. Biol. 2022, 11, 343–352.
- Baumschlager, A.; Aoki, S.K.; Khammash, M. Dynamic blue light-inducible T7 RNA polymerases (Opto-T7RNAPs) for precise spatiotemporal gene expression control. ACS Synth. Biol. 2017, 6, 2157–2167.
- Rouches, M.V.; Xu, Y.; Cortes, L.B.G.; Lambert, G. A plasmid system with tunable copy number. Nat. Commun. 2022, 13, 3908.
- Ikemura, T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1981, 146, 1–21.
- Boël, G.; Letso, R.; Neely, H.; Price, W.N.; Wong, K.H.; Su, M.; Luff, J.D.; Valecha, M.; Everett, J.K.; Acton, T.B.; et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature 2016, 529, 358–363.
- Fuhrmann, M.; Hausherr, A.; Ferbitz, L.; Schödl, T.; Heitzer, M.; Hegemann, P. Monitoring dynamic expression of nuclear genes in Chlamydomonas reinhardtii by using a synthetic luciferase reporter gene. Plant Mol. Biol. 2004, 55, 869–881.
- Kleber-Janke, T.; Becker, W.M. Use of modified BL21(DE3) Escherichia coli cells for high-level expression of recombinant peanut allergens affected by poor codon usage. Protein Expr. Purif. 2000, 19, 419–424.
- Novy, R.; Drott, D.; Yaeger, K.; Mierendorf, R. Overcoming the codon bias of E. coli for enhanced protein expression. Innovations 2001, 12, 1–3.
- Komar, A.A. The Yin and Yang of codon usage. Hum. Mol. Genet. 2016, 25, R77–R85.
- Chemla, Y.; Peeri, M.; Heltberg, M.L.; Eichler, J.; Jensen, M.H.; Tuller, T.; Alfonta, L. A possible universal role for mRNA secondary structure in bacterial translation revealed using a synthetic operon. Nat. Commun. 2020, 11, 4827.
- Lenz, G.; Doron-Faigenboim, A.; Ron, E.Z.; Tuller, T.; Gophna, U. Sequence Features of E. coli mRNAs Affect Their Degradation. PLoS ONE 2011, 6, e28544.
- Siller, E.; DeZwaan, D.C.; Anderson, J.F.; Freeman, B.C.; Barral, J.M. Slowing Bacterial Translation Speed Enhances Eukaryotic Protein Folding Efficiency. J. Mol. Biol. 2010, 396, 1310–1318.
- Angov, E.; Hillier, C.J.; Kincaid, R.L.; Lyon, J.A. Heterologous Protein Expression Is Enhanced by Harmonizing the Codon Usage Frequencies of the Target Gene with those of the Expression Host. PLoS ONE 2008, 3, e2189.
- Gasser, B.; Saloheimo, M.; Rinas, U.; Dragosits, M.; Rodríguez-Carmona, E.; Baumann, K.; Giuliani, M.; Parrilli, E.; Branduardi, P.; Lang, C.; et al. Protein folding and conformational stress in microbial cells producing recombinant proteins: A host comparative overview. Microb. Cell Fact. 2008, 7, 11.
- Goemans, C.; Denoncin, K.; Collet, J.F. Folding mechanisms of periplasmic proteins. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 1517–1528.
- Skerra, A.; Pluckthun, A. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science 1988, 240, 1038–1041.
- Chen, C.; Snedecor, B.; Nishihara, J.C.; Joly, J.C.; McFarland, N.; Andersen, D.C.; Battersby, J.E.; Champion, K.M. High-level accumulation of a recombinant antibody fragment in the periplasm of Escherichia coli requires a triple-mutant (degP prc spr) host strain. Biotechnol. Bioeng. 2004, 85, 463–474.
- Carter, P.; Kelley, R.F.; Rodrigues, M.L.; Snedecor, B.; Covarrubias, M.; Velligan, M.D.; Wong, W.L.T.; Rowland, A.M.; Kotts, C.E.; Carver, M.E.; et al. High level Escherichia coli expression and production of a bivalent humanized antibody fragment. Biotechnology 1992, 10, 163–167.
- Huang, M.N.; Lu, X.Y.; Zong, H.; Bin, Z.G.; Shen, W. Bioproduction of trans-10, cis-12-Conjugated Linoleic Acid by a Highly Soluble and Conveniently Extracted Linoleic Acid Isomerase and an Extracellularly Expressed Lipase from Recombinant Escherichia coli Strains. J. Microbiol. Biotechnol. 2018, 28, 739–747.
More
Information
Subjects:
Medicine, Research & Experimental
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
773
Revisions:
2 times
(View History)
Update Date:
08 Dec 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




