
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ilya Kirov | -- | 1724 | 2023-12-05 15:49:34 | | | |
| 2 | Jessie Wu | + 3 word(s) | 1727 | 2023-12-06 06:21:47 | | | | |
| 3 | Jessie Wu | Meta information modification | 1727 | 2023-12-06 06:22:16 | | |
Video Upload Options
Genetic diversity is a key factor for plant breeding. The birth of novel genic and genomic variants is also crucial for plant adaptation in nature. Therefore, the genomes of almost all living organisms possess natural mutagenic mechanisms. Transposable elements (TEs) are a major mutagenic force driving genetic diversity in wild plants and modern crops. The relatively rare TE transposition activity during the thousand-year crop domestication process has led to the phenotypic diversity of many cultivated species. The utilization of TE mutagenesis by artificial and transient acceleration of their activity in a controlled mode is an attractive foundation for a novel type of mutagenesis called TE-mediated biological mutagenesis.
1. Introduction
2. Heterologous Expression of Transposable Elements via Plant Transformation
3. Implication of DNA-Methylation-Deficient Genotypes
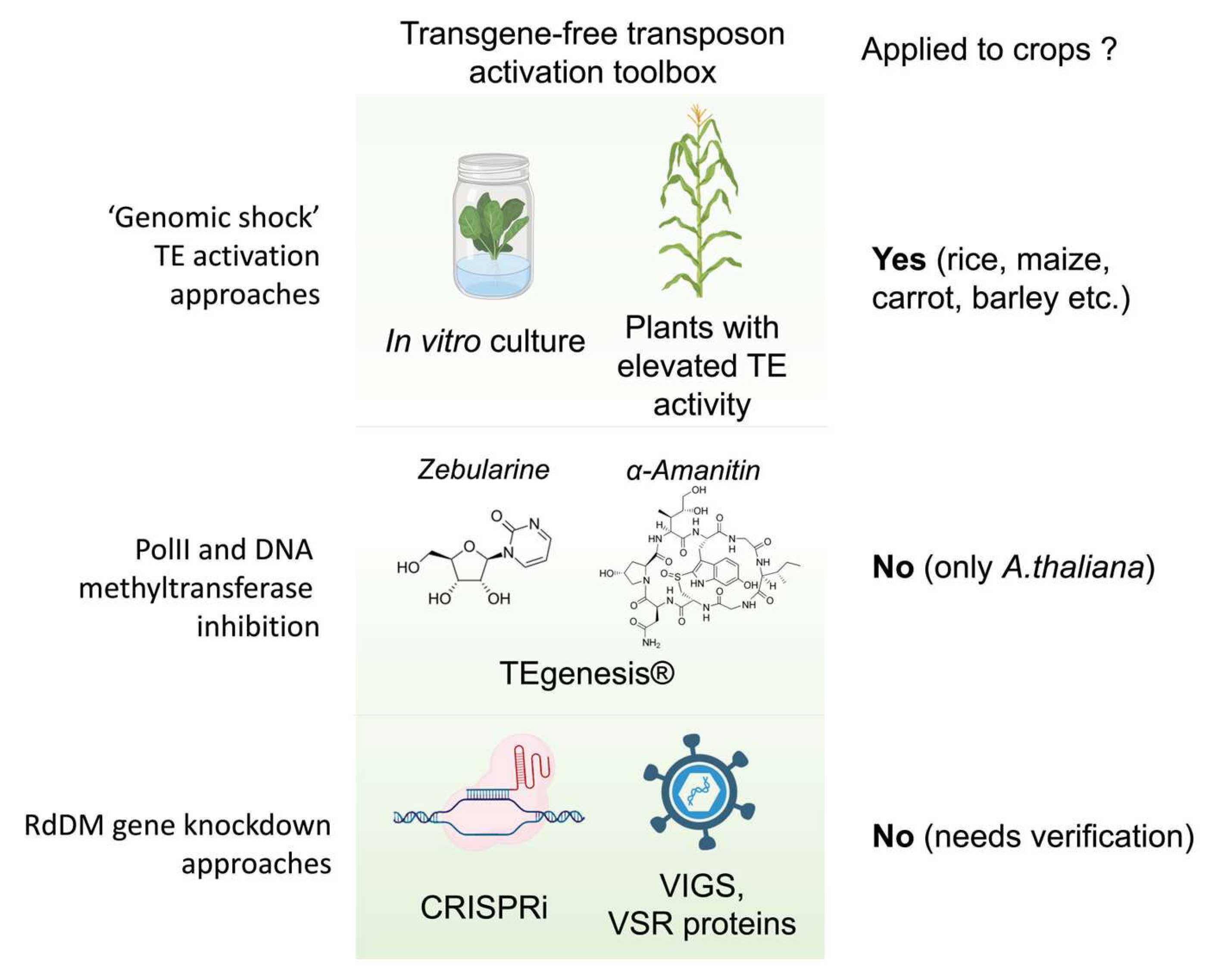
4. Utilization of Mutagenic Plant Lines with Reactivated Transposable Elements
References
- Ma, L.; Kong, F.; Sun, K.; Wang, T.; Guo, T. From Classical Radiation to Modern Radiation: Past, Present, and Future of Radiation Mutation Breeding. Front. Public Health 2021, 9, 768071.
- Greenwood, J.R.; Zhang, X.; Rathjen, J.P. Precision Genome Editing of Crops for Improved Disease Resistance. Curr. Biol. 2023, 33, R650–R657.
- Zhang, Y.; Massel, K.; Godwin, I.D.; Gao, C. Applications and Potential of Genome Editing in Crop Improvement. Genome Biol. 2018, 19, 210.
- Belfield, E.J.; Brown, C.; Ding, Z.J.; Chapman, L.; Luo, M.; Hinde, E.; Es, S.W.; van Johnson, S.; Ning, Y.; Zheng, S.J.; et al. Thermal Stress Accelerates Arabidopsis Thaliana Mutation Rate. Genome Res. 2021, 31, 40–50.
- Lu, Z.; Cui, J.; Wang, L.; Teng, N.; Zhang, S.; Lam, H.-M.; Zhu, Y.; Xiao, S.; Ke, W.; Lin, J.; et al. Genome-Wide DNA Mutations in Arabidopsis Plants after Multigenerational Exposure to High Temperatures. Genome Biol. 2021, 22, 160.
- Baduel, P.; Leduque, B.; Ignace, A.; Gy, I.; Gil, J.; Loudet, O.; Colot, V.; Quadrana, L. Genetic and Environmental Modulation of Transposition Shapes the Evolutionary Potential of Arabidopsis Thaliana. Genome Biol 2021, 22, 138.
- Hämälä, T.; Ning, W.; Kuittinen, H.; Aryamanesh, N.; Savolainen, O. Environmental Response in Gene Expression and DNA Methylation Reveals Factors Influencing the Adaptive Potential of Arabidopsis Lyrata. eLife 2022, 11, e83115.
- Quadrana, L.; Silveira, A.B.; Mayhew, G.F.; LeBlanc, C.; Martienssen, R.A.; Jeddeloh, J.A.; Colot, V. The Arabidopsis Thaliana Mobilome and Its Impact at the Species Level. eLife 2016, 5, e15716.
- Merkulov, P.; Gvaramiya, S.; Dudnikov, M.; Komakhin, R.; Omarov, M.; Kocheshkova, A.; Konstantinov, Z.; Soloviev, A.; Karlov, G.; Divashuk, M.; et al. Cas9-targeted Nanopore Sequencing Rapidly Elucidates the Transposition Preferences and DNA Methylation Profiles of Mobile Elements in Plants. J. Integr. Plant Biol. 2023, 65, 2242–2261.
- Quadrana, L.; Etcheverry, M.; Gilly, A.; Caillieux, E.; Madoui, M.-A.; Guy, J.; Silveira, A.B.; Engelen, S.; Baillet, V.; Wincker, P.; et al. Transposition Favors the Generation of Large Effect Mutations That May Facilitate Rapid Adaption. Nat. Commun. 2019, 10, 3421.
- Paszkowski, J. Controlled Activation of Retrotransposition for Plant Breeding. Curr. Opin. Biotechnol. 2015, 32, 200–206.
- Dubin, M.J.; Scheid, O.M.; Becker, C. Transposons: A Blessing Curse. Curr. Opin. Plant Biol. 2018, 42, 23–29.
- Domínguez, M.; Dugas, E.; Benchouaia, M.; Leduque, B.; Jiménez-Gómez, J.M.; Colot, V.; Quadrana, L. The Impact of Transposable Elements on Tomato Diversity. Nat. Commun. 2020, 11, 4058.
- Cai, X.; Lin, R.; Liang, J.; King, G.J.; Wu, J.; Wang, X. Transposable Element Insertion: A Hidden Major Source of Domesticated Phenotypic Variation in Brassica Rapa. Plant Biotechnol. J. 2022, 20, 1298–1310.
- Studer, A.; Zhao, Q.; Ross-Ibarra, J.; Doebley, J. Identification of a Functional Transposon Insertion in the Maize Domestication Gene Tb1. Nat. Genet. 2011, 43, 1160–1163.
- Wei, L.; Cao, X. The Effect of Transposable Elements on Phenotypic Variation: Insights from Plants to Humans. Sci. China Life Sci. 2016, 59, 24–37.
- Lisch, D. How Important Are Transposons for Plant Evolution? Nat. Rev. Genet. 2013, 14, 49–61.
- Latzel, V.; Puy, J.; Thieme, M.; Bucher, E.; Götzenberger, L.; Bello, F. Phenotypic Diversity Influenced by a Transposable Element Increases Productivity and Resistance to Competitors in Plant Populations. J. Ecol. 2023, 111, 2376–2387.
- Lieberman-Lazarovich, M.; Kaiserli, E.; Bucher, E.; Mladenov, V. Natural and Induced Epigenetic Variation for Crop Improvement. Curr. Opin. Plant Biol. 2022, 70, 102297.
- Thieme, M.; Lanciano, S.; Balzergue, S.; Daccord, N.; Mirouze, M.; Bucher, E. Inhibition of RNA Polymerase II Allows Controlled Mobilisation of Retrotransposons for Plant Breeding. Genome Biol. 2017, 18, 134.
- Thieme, M.; Brêchet, A.; Bourgeois, Y.; Keller, B.; Bucher, E.; Roulin, A.C. Experimentally Heat-induced Transposition Increases Drought Tolerance in Arabidopsis Thaliana. New Phytol. 2022, 236, 182–194.
- Roquis, D.; Robertson, M.; Yu, L.; Thieme, M.; Julkowska, M.; Bucher, E. Genomic Impact of Stress-Induced Transposable Element Mobility in Arabidopsis. Nucleic Acids Res. 2021, 49, gkab828.
- Fedoroff, N.; Wessler, S.; Shure, M. Isolation of the Transposable Maize Controlling Elements Ac and Ds. Cell 1983, 35, 235–242.
- Baker, B.; Schell, J.; Lörz, H.; Fedoroff, N. Transposition of the Maize Controlling Element “Activator” in Tobacco. Proc. Natl. Acad. Sci. USA 1986, 83, 4844–4848.
- Yamazaki, M.; Tsugawa, H.; Miyao, A.; Yano, M.; Wu, J.; Yamamoto, S.; Matsumoto, T.; Sasaki, T.; Hirochika, H. The Rice Retrotransposon Tos17 Prefers Low-Copy-Number Sequences as Integration Targets. Mol. Genet. Genom. 2001, 265, 336–344.
- Okamoto, H.; Hirochika, H. Efficient Insertion Mutagenesis of Arabidopsis by Tissue Culture-induced Activation of the Tobacco Retrotransposon Tto1. Plant J. 2000, 23, 291–304.
- Courtial, B.; Feuerbach, F.; Eberhard, S.; Rohmer, L.; Chiapello, H.; Camilleri, C.; Lucas, H. Tnt1 Transposition Events Are Induced by in Vitro Transformation of Arabidopsis Thaliana, and Transposed Copies Integrate into Genes. Mol. Genet. Genom. 2001, 265, 32–42.
- D’Erfurth, I.; Cosson, V.; Eschstruth, A.; Lucas, H.; Kondorosi, A.; Ratet, P. Efficient Transposition of the Tnt1 Tobacco Retrotransposon in the Model Legume Medicago Truncatula. Plant J. 2003, 34, 95–106.
- Kim, E.Y.; Wang, L.; Lei, Z.; Li, H.; Fan, W.; Cho, J. Ribosome Stalling and SGS3 Phase Separation Prime the Epigenetic Silencing of Transposons. Nat. Plants 2021, 7, 303–309.
- Nicolau, M.; Picault, N.; Moissiard, G. The Evolutionary Volte-Face of Transposable Elements: From Harmful Jumping Genes to Major Drivers of Genetic Innovation. Cells 2021, 10, 2952.
- Liu, B.; Zhao, M. How Transposable Elements Are Recognized and Epigenetically Silenced in Plants? Curr. Opin. Plant Biol. 2023, 75, 102428.
- Liu, P.; Cuerda-Gil, D.; Shahid, S.; Slotkin, R.K. The Epigenetic Control of the Transposable Element Life Cycle in Plant Genomes and Beyond. Annu. Rev. Genet. 2022, 56, 63–87.
- Panda, K.; Ji, L.; Neumann, D.A.; Daron, J.; Schmitz, R.J.; Slotkin, R.K. Full-Length Autonomous Transposable Elements Are Preferentially Targeted by Expression-Dependent Forms of RNA-Directed DNA Methylation. Genome Biol. 2016, 17, 170.
- Catoni, M.; Cortijo, S. Chapter Four EpiRILs Lessons From Arabidopsis. Adv. Bot. Res. 2018, 88, 87–116.
- Kooke, R.; Johannes, F.; Wardenaar, R.; Becker, F.; Etcheverry, M.; Colot, V.; Vreugdenhil, D.; Keurentjes, J.J.B. Epigenetic Basis of Morphological Variation and Phenotypic Plasticity in Arabidopsis Thaliana. Plant Cell 2015, 27, 337–348.
- Reinders, J.; Wulff, B.B.H.; Mirouze, M.; Marí-Ordóñez, A.; Dapp, M.; Rozhon, W.; Bucher, E.; Theiler, G.; Paszkowski, J. Compromised Stability of DNA Methylation and Transposon Immobilization in Mosaic Arabidopsis Epigenomes. Genes Dev. 2009, 23, 939–950.
- Johannes, F.; Porcher, E.; Teixeira, F.K.; Saliba-Colombani, V.; Simon, M.; Agier, N.; Bulski, A.; Albuisson, J.; Heredia, F.; Audigier, P.; et al. Assessing the Impact of Transgenerational Epigenetic Variation on Complex Traits. PLoS Genet. 2009, 5, e1000530.
- Tsukahara, S.; Kobayashi, A.; Kawabe, A.; Mathieu, O.; Miura, A.; Kakutani, T. Bursts of Retrotransposition Reproduced in Arabidopsis. Nature 2009, 461, 423–426.
- Mirouze, M.; Reinders, J.; Bucher, E.; Nishimura, T.; Schneeberger, K.; Ossowski, S.; Cao, J.; Weigel, D.; Paszkowski, J.; Mathieu, O. Selective Epigenetic Control of Retrotransposition in Arabidopsis. Nature 2009, 461, 427–430.
- Robertson, D.S. Characterization of a Mutator System in Maize. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1978, 51, 21–28.
- McClintock, B. The Significance of Responses of the Genome to Challenge. Science 1984, 226, 792–801.
- Richardson, A.E.; Hake, S. The Power of Classic Maize Mutants: Driving Forward Our Fundamental Understanding of Plants. Plant Cell 2021, 34, 2505–2517.
- Lisch, D. Mutator Transposons. Trends Plant Sci. 2002, 7, 498–504.
- Sasaki, T.; Kato, K.; Hosaka, A.; Fu, Y.; Toyoda, A.; Fujiyama, A.; Tarutani, Y.; Kakutani, T. Arms Race between Anti-silencing and RdDM in Noncoding Regions of Transposable Elements. EMBO Rep. 2023, 24, e56678.
- Barker, R.F.; Thompson, D.V.; Talbot, D.R.; Swanson, J.; Bennetzen, J.L. Nucleotide Sequence of the Maize Transposable Element Mul. Nucleic Acids Res. 1984, 12, 6924.
- Alleman, M.; Freeling, M. The Mu Transposable Elements Of Maize: Evidence For Transposition And Copy Number Regulation During Development. Genetics 1986, 112, 107–119.
- Slotkin, R.K.; Freeling, M.; Lisch, D. Mu Killer Causes the Heritable Inactivation of the Mutator Family of Transposable Elements in Zea Mays. Genetics 2003, 165, 781–797.
- Burgess, D.; Li, H.; Zhao, M.; Kim, S.Y.; Lisch, D. Silencing of Mutator Elements in Maize Involves Distinct Populations of Small RNAs and Distinct Patterns of DNA Methylation. Genetics 2020, 215, 379–391.
- Kolkman, J.M.; Conrad, L.J.; Farmer, P.R.; Hardeman, K.; Ahern, K.R.; Lewis, P.E.; Sawers, R.J.H.; Lebejko, S.; Chomet, P.; Brutnell, T.P. Distribution of Activator (Ac) Throughout the Maize Genome for Use in Regional Mutagenesis. Genetics 2005, 169, 981–995.
- Bai, L.; Singh, M.; Pitt, L.; Sweeney, M.; Brutnell, T.P. Generating Novel Allelic Variation Through Activator Insertional Mutagenesis in Maize. Genetics 2007, 175, 981–992.
- Bensen, R.J.; Johal, G.S.; Crane, V.C.; Tossberg, J.T.; Schnable, P.S.; Meeley, R.B.; Briggs, S.P. Cloning and Characterization of the Maize An1 Gene. Plant Cell 1995, 7, 75–84.
- Naito, K.; Cho, E.; Yang, G.; Campbell, M.A.; Yano, K.; Okumoto, Y.; Tanisaka, T.; Wessler, S.R. Dramatic Amplification of a Rice Transposable Element during Recent Domestication. Proc. Natl. Acad. Sci. USA 2006, 103, 17620–17625.
- Simmons, C.R.; Lafitte, H.R.; Reimann, K.S.; Brugière, N.; Roesler, K.; Albertsen, M.C.; Greene, T.W.; Habben, J.E. Successes and Insights of an Industry Biotech Program to Enhance Maize Agronomic Traits. Plant Sci. 2021, 307, 110899.
- Yasuda, K.; Ito, M.; Sugita, T.; Tsukiyama, T.; Saito, H.; Naito, K.; Teraishi, M.; Tanisaka, T.; Okumoto, Y. Utilization of Transposable Element MPing as a Novel Genetic Tool for Modification of the Stress Response in Rice. Mol. Breed. 2013, 32, 505–516.





