1. Introduction
The main factor in the selection of a cancer treatment regimen is based on the place of tumor formation and its stage. Recent research has demonstrated that the molecular genetics of cancer also play a crucial role in the determination of effective treatment
[1]. The diagnosis of genetic disorders allows for the identification of genes with an increased or decreased expression level. The use of drugs targeted at specific molecular targets may increase the effectiveness of treatment as well as reduce the risk of side effects associated with traditional chemotherapy
[2]. One of the techniques used in targeted therapy is aptamers. Aptamers may be promising anti-cancer therapeutics due to their ease of modification and functionalization
[3]. Due to the mechanism of action, aptamers can be divided into three groups: antagonists (blocking protein-protein interactions, e.g., AS1411 blocking nucleolin, AP-50 inhibiting NF-kB)
[4], agonists (stimulating receptor activation), and bi-specific aptamers (e.g., MRP1-CD28, which inhibits CTLA-4 and stimulates CD28)
[5]. Additionally, aptamers can also be used as carriers of chemotherapeutic agents to deliver them to target tissues (e.g., doxorubicin)
[2].
2. Potential New Target for Aptamer and Aptamer Chimera Development for the Treatment of OC
The potential applications of aptamers in the diagnosis and treatment of tumors, particularly in the context of OC, offer substantial opportunities. Simultaneously, there is a need to identify additional OC biomarkers apart from those currently known and to develop aptamer-based probes tailored for detecting proteins present in low quantities or for facilitating in vivo imaging. These advancements are crucial for enabling early-stage diagnosis of OC
[6]. Moreover, given the relatively small molecular size of aptamers, it is imperative to explore suitable materials that can reduce their clearance rate within the body. Combining aptamers with anti-tumor drugs presents an avenue to enhance therapeutic efficacy while mitigating adverse effects. Additionally, the identification of valuable gene targets is essential, and further research should investigate nanomaterials loaded with aptamers, miRNAs, LNAs (locked nucleic acids), and siRNAs to modulate the expression of specific target genes. The ultimate goal is to design aptamer/drug systems that exhibit high specificity for tumor tissues in vivo while minimizing distribution in other tissues. As of now, several aptamers have been documented for their ability to target proteins associated with OC, including aptamers targeting MUC1, CA125, HE4, CD44, and programmed death-ligand 1 (PD-L1)
[7][8][9][10][11].
Simaeys et al. have reported the discovery of a set of aptamers capable of specifically recognizing the ovarian clear-cell carcinoma (OCCC) subtype TOV-21 G
[12]. Subsequently, they identified stress-induced phosphoprotein 1 (STIP1) as one of the target proteins, indicating its potential utility as a biomarker for OC
[13]. The pursuit of identifying novel targets for the development of aptamers and aptamer chimeras as potential therapeutic agents in OC requires a comprehensive understanding of the disease and the intricate molecular pathways involved. To this end, Lee and collaborators have conducted an extensive multi-omics analysis characterized by high-depth scrutiny to discern disparities in the molecular and cellular attributes of highly clinically annotated samples of HGSC originating from both primary and multiple metastatic sites
[14]. They observed a notable increase in the loss of copies of the
NF1 gene, as well as reduced levels of its RNA and protein products, fewer cytotoxic T-lymphocyte precursors (CTLPs), and a significantly higher presence of strongly binding neoantigens in the group of patients who underwent primary complete gross resection (R0) compared to those who received neoadjuvant chemotherapy (NACT). Furthermore, the R0 group exhibited a significant rise in T-cell infiltration and a decrease in macrophage numbers. Transcriptomic and proteomic differences were identified in this group
[14][15]. Despite differences in copy number variation and T-cell infiltration between the two groups, these disparities were not as extensive as anticipated. These findings suggest that critical molecular distinctions between such tumors cannot be comprehensively unveiled through bulk molecular analyses. Consequently, the researchers conducted an extensive spatial examination of both the epithelial and stromal components within tumor tissue to investigate the heterogeneity of HGSC and explore how the dynamic interactions between the tumor and its microenvironment vary between individuals who respond well and poorly to NACT. To obtain these insights, they conducted an in-depth spatial transcriptomic profiling directly within the tissue
[16][17].
In a comprehensive multiomics examination of tumor profiles, Stur et al. compared patients with HGSC who survived for a minimum of 10 years with those who survived for only 3 years. Their investigation identified
TMEM62 as a potential therapeutic target to enhance survival. The overexpression of
TMEM62 was distinctive in the first group of tumors but not observed in the second group, and this increased expression was associated with alterations in cell survival pathways, including upregulation of senescence markers. Furthermore, they observed that the
MAL gene exhibited the highest upregulation in patients with short-term survival. These findings suggest that the restoration of
TMEM62 could represent a novel approach to the treatment of HGSC. Importantly, these discoveries may hold implications for the development of biomarkers and intervention strategies aimed at improving patient outcomes
[17]. Two pivotal discoveries emerged from their investigations: first, they underscored the significance of the stromal component as a potential driver of suboptimal responses to chemotherapy, and second, they identified distinct clusters (in terms of their distribution and composition) in tissues from patients who responded well to NACT compared to those who responded poorly, both at a collective and individual patient level. These clusters could only be discerned through the in situ technique. Notably, when focusing on the epithelial-mesenchymal transition pathway, they observed substantial variations in its upregulation or downregulation among different regions within the same tumor tissue, as well as among specific areas in the tissues across the entire patient cohort. This observation suggests that particular cell populations may contribute to the inadequate response to therapy
[18].
In a separate study, Ye et al. conducted an integrative genomic and transcriptomic profiling aimed at identifying molecular subtypes and prognostic markers in OCCC, with a specific focus on immune-related pathways. Through this integrated analysis, they delineated two distinct OCCC molecular subtypes with differing functional characteristics and prognoses. Notably, the immune subset exhibited enrichment in PD-1 and PI3K-AKT-mTOR signaling pathways. Leveraging their data, the researchers constructed a robust prognostic immune signature for OCCC patients, which was not only derived from their dataset but also validated in publicly available repositories. The potential of the immune/non-immune classification as a predictive tool for targeted therapy warrants further investigation through prospective studies
[19].
Wang et al. conducted a comprehensive multiomics investigation focused on OC, revealing a notable degree of tumor heterogeneity within this disease. They acquired high-resolution profiles of HGSC, meticulously dissecting both the intertumor and intratumor heterogeneities. This effort led to the identification of numerous potentially crucial variations occurring during tumorigenesis and the elucidation of regulatory networks within HGSC. Through utilizing integrated multiomics analyses, it was revealed that the increased activity of interferon signaling and metallothioneins stemmed from a combination of demethylation in their promoters and hypomethylation in satellite regions and LINE1. Furthermore, the investigation uncovered potential critical transcription factors that govern glycolysis by leveraging chromatin accessibility data. Interestingly, observed patterns in gene expression and DNA methylation remained consistent in both primary and abdominal metastatic tumor cells of matched genetic lineage, suggesting that metastatic cells may potentially exist as subclones within primary tumors. Notably, cancer cell lineages exhibiting heightened residual DNA methylation levels, along with elevated expression of CCN1 and HSP90AA1, displayed a heightened propensity for metastasis.
Guo et al. integrated single-cell sequencing data from 12 OC patients to construct a comprehensive cell atlas containing normal epithelium, primary carcinoma, and metastatic carcinoma states. Within this atlas, they pinpointed an upregulated gene called RAB13, which had not previously been associated with OC. Subsequently, they conducted experiments to validate RAB13’s pro-metastatic effects on OC cell lines, demonstrating its promotion of cell migration and invasion in vitro. Further investigations delved into the clinical implications of RAB13’s expression levels using datasets from the Cancer Genome Atlas (TCGA), revealing a significant association between RAB13 expression and adverse prognosis and tumor progression. Additionally, the study employed predictive methods to identify two cytoskeleton inhibitors that could potentially target RAB13.
Nonetheless, a significant challenge in the application of specific aptamers into future clinical therapeutics for cancer treatment results from the limited number of clinical trials dedicated to exploring these agents. Furthermore, the relatively small number of potential molecular targets for aptamers represents another problem. To overcome these challenges, there is a need to increase clinical trials for aptamer therapies in various cancers. Additionally, efforts should be directed towards identifying and characterizing a wider range of molecular targets for aptamers.
3. Aptmers for the Treatment of OC
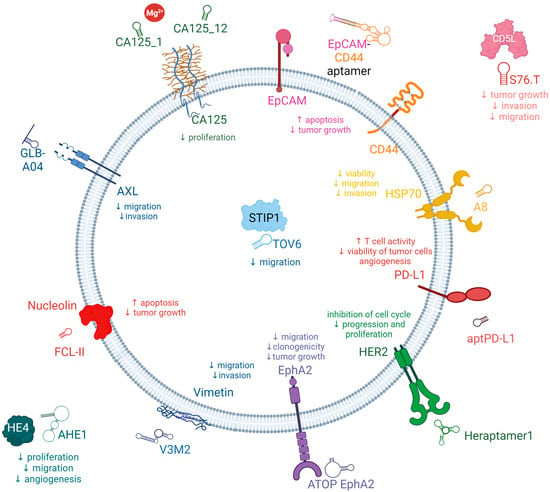
Here, here summarized the latest aptamer reports in the literature (Figure 1), in which aptamers specifically interact with biomarkers associated with poor patient survival in OC.
Figure 1. Schema of the aptamers and targets developed for the treatment of OC.
3.1. AXL Receptor Tyrosine Kinase (AXL)
Receptor tyrosine kinases (RTKs) are prominent therapeutic targets due to their pivotal involvement in regulating growth factor signaling and the metastatic behavior of tumors. AXL, a member of the receptor tyrosine kinase family, belongs to the TAM (Tyro3-AXL-Mer) receptor kinase subfamily along with its homologs, Tyro3 and Mer
[20][21]. AXL has been identified as an oncogenic factor due to its ability to enhance cancer cell survival, proliferation, invasion, and metastasis. The primary ligand responsible for activating AXL is growth-arrest-specific protein 6 (Gas6), a γ-carboxylate protein that exhibits a strong affinity for the AXL receptor. Upon binding, Gas6 induces dimerization and autophosphorylation of specific tyrosine residues in the AXL receptor. Consequently, this initiates the recruitment, phosphorylation, and subsequent activation of multiple signaling pathways, including PI3K, MAPK, and PKC
[22]. AXL exhibits widespread expression and has been detected in various organs and cell types, including monocytes, macrophages, and endothelial cells present in the heart, skeletal muscles, liver, and kidneys. The dysregulation of the Gas6/AXL interaction, either through overexpression or heightened activity, has been documented in different cancer types, including prostate, esophageal, thyroid, breast, lung, liver, OC, and glioblastoma, and is associated with poor prognosis. Moreover, this dysregulation has been correlated with unfavorable prognostic outcomes
[20][21][23][24][25]. In a study conducted by Rankin et al., it was demonstrated that AXL exhibits elevated expression levels specifically in high-grade serous ovarian tumors and metastatic ovarian tumors
[26]. However, normal ovarian epithelium and tumor stroma do not display significant AXL expression. Considering the association of AXL with prognostic indications in OC, targeting AXL in ovarian tumor cells presents a promising therapeutic approach with the potential to inhibit cancer progression. The down-modulation of AXL expression would result in a decrease in OC tumor growth and an improvement in patients’ overall survival
[20][24].
Various strategies can be employed to modify aptamers, aiming to extend their half-life in serum. These modifications can be incorporated directly during the SELEX process or through post-SELEX optimizations. Given that wild-type aptamer molecules possess a notably short half-life, primarily due to their rapid clearance by the kidneys and susceptibility to nuclease-mediated degradation, their utility in physiological conditions is constrained. Consequently, a range of biochemistry-based approaches have been employed to engineer modified aptamers aimed at addressing this instability issue while optimizing the pharmacokinetic and pharmacodynamic attributes of aptamers. These specific modifications enable the targeted delivery of aptamers to precise cellular locations. These modifications involve functional optimization, multimerization, or truncation, all of which have demonstrated enhanced stability and binding efficacy. The field of aptamer technology encompasses a diverse array of modification designs, conjugation strategies, and linkage methodologies. Chemical modifications of aptamers are a common strategy utilized to bolster their stability and functionality. Notably, hydrophobic fatty acids and amphiphilic PEG components have been commonly used as agents to enhance the longevity of aptamers in drug candidates. However, it is essential to consider that high-molecular-mass PEG components have a substantial molecular weight proportion, posing a significant challenge when attempting to increase subcutaneous dosages to maximize therapeutic potential. Additionally, it is worth noting that clinical studies have reported serious or even fatal immune responses related to PEG moieties. Therefore, there are safety concerns associated with the use of high-molecular-mass PEG moieties in aptamer modifications Notably, hydrophobic fatty acids and amphiphilic PEG components have been commonly used as agents to enhance the longevity of aptamers in drug candidates. However, it is essential to consider that high-molecular-mass PEG components have a substantial molecular weight proportion, posing a significant challenge when attempting to increase subcutaneous dosages to maximize therapeutic potential. Additionally, it is worth noting that clinical studies have reported serious or even fatal immune responses related to PEG moieties. Therefore, there are safety concerns associated with the use of high-molecular-mass PEG moieties in aptamer modifications
[27][28].
Kanlikilicer et al. developed a nuclease-resistant AXL aptamer to specifically target phosphor-AXL-RTK and assessed its efficacy as an antitumor agent using various intraperitoneal injection animal models. In combination with paclitaxel, the AXL aptamer demonstrated a notable augmentation of the antitumor effectiveness of paclitaxel chemotherapy. Consequently, the synergistic application of AXL-APTAMER in conjunction with chemotherapy exhibits promising potential as a strategy for addressing drug-resistant OC. Moreover, it offers the advantage of targeted delivery of chemotherapy specifically to tumor cells, thereby minimizing the adverse effects of chemotherapy on healthy cells and reducing chemotherapy-induced damage. Kanlikilicer et al. demonstrated the effectiveness of chemical modifications applied to aptamers, addressing inherent limitations and offering an attractive and efficient solution. Data gathered by Kalikilicer et al. highlight the successful application of 2′-fluoro and monothiophosphate modifications, which rendered the AXL-DNA-APTAMER stable for up to 24 h in high concentrations of human serum. This stability allowed for the effective inhibition of AXL phosphorylation, consequently leading to robust and prolonged suppression of downstream oncogenic signaling pathways, including those related to cancer metastasis and survival
[23].
Amero et al. devised an innovative approach to enhance the stability and bioavailability of aptamers by converting RNA aptamers into modified DNA aptamers that specifically target phospho-AXL. Through a comprehensive analysis of a library containing 17 converted modified DNA aptamers, they identified two optimal aptamer candidates, namely GLB-G25 and GLB-A04, which exhibited superior bioavailability, and stability, and demonstrated potent antitumor effects in in vitro experiments. Notably, the incorporation of backbone modifications such as thiophosphate or dithiophosphate, along with covalent modification of the 5′-end of the aptamer using polyethylene glycol, resulted in optimized pharmacokinetic properties. These modifications improved the in vivo stability of the aptamers by reducing nuclease hydrolysis and renal clearance, ultimately leading to sustained and potent inhibition of AXL even at very low doses. Administration of these modified aptamers in orthotopic mouse models of OC yielded substantial reductions in tumor growth and the incidence of metastasis. This remarkable inhibition of the phospho-AXL target exemplifies the potential to enhance the specificity and bioavailability of aptamers through chemical modifications. Consequently, these findings establish a solid basis for further translational studies aiming to bring these aptamer candidates and companion biomarkers into clinical practice
[20].
3.2. Cancer Antigen 125 (CA125)
Cancer antigen 125 (CA125), also known as MUC16, is a transmembrane mucin with a substantial number of attached sugar molecules. It has a large molecular weight and is found to be excessively produced in 80% of cases of OC
[29]. The concentration of CA125 in the bloodstream is routinely measured to aid in the diagnosis of OC, assess the effectiveness of treatment, and monitor disease progression
[30]. Significantly elevated levels of CA125 are detected in 50% of patients at the early stage of the disease and in 85% of patients with advanced OC, establishing it as the widely accepted biomarker for OC in clinical practice
[31][32][33]. The CA125 antigen is typically distributed throughout various anatomical sites in the human body. It is detected in the cervical mucus of healthy women and is likely synthesized and released by endocervical cells. The amniotic fluid and chorionic membrane of the developing fetus contain abundant quantities of CA125. Additionally, CA125 expression is observed in human milk, epithelial cells lining the airways, respiratory glands, and bronchial mucus. Furthermore, CA125 is expressed in diverse tissues, including the endocervix, endometrium, pleura, pericardium, peritoneum, secretory mammary glands, apocrine sweat glands, intestines, lungs, and kidneys. MUC16 (CA125) actively participates in the development of ovarian tumors and the formation of metastases by binding to the peritoneal mesothelium. In addition to its involvement in metastasis, MUC16 hinders the destruction of tumor cells by NK cells. Moreover, MUC16 obstructs the recognition of cancer cells by NK cells, consequently promoting the survival of these cells
[29][31][32][33][34].
Scoville et al. employed the One-Pot SELEX method to select two aptamers, CA125_1 and CA125_12, both generated without the use of primers. These aptamers demonstrated the ability to bind to clinically relevant concentrations of the target protein. Interestingly, the presence of Mg
2+ ions exerted distinct effects on the binding behavior of the aptamers. Specifically, the binding of CA125_1 required the presence of Mg
2+ ions, whereas the presence of these ions abolished the binding of CA125_12. In summary, the One-Pot SELEX approach proved to be a promising selection method, resulting in the identification of DNA aptamers targeting a protein of significant clinical importance
[33].
Lamberti et al. successfully isolated two RNA aptamers, CA125.1 and CA125.11, specifically targeting the CA125 tumor marker. The anti-CA125 aptamers were obtained using a protein/SELEX strategy, involving the incubation of a library with purified His-tagged CA125 protein. The transcription process was carried out in the presence of 20-F-Py, ATP, GTP, dithiothreitol, RNase inhibitors, inorganic pyrophosphatase, and a modified variant of T7 RNA polymerase, employed specifically to enhance yield. In the process, 20F-Py RNAs were employed to increase the aptamers’ resistance against degradation by nucleases. RT-qPCR analysis demonstrated that CA125.1 exhibited significantly superior binding properties compared to CA125.11. This finding was further confirmed through surface plasmon resonance (SPR) binding experiments, where the ligand-protein was immobilized on a chip, and both aptamers were tested for their response in terms of response units (RU). Further investigation focused on CA125.1 as the analyte in solution, leading to the determination of initial kinetic parameters. These parameters indicated a dissociation constant in the nanomolar range. This outcome suggests that the selected aptamer, CA125.1, holds promise as a potential diagnostic tool, particularly as a bioreceptor in SPR-based biosensors, warranting further development
[34].
Gedi et al. presented a method for selecting an anti-CA125 aptamer and its subsequent utilization in a biochip featuring a three-dimensional network of carbon nanotubes (3DN-CNTs) for CA125 detection. The aptamer, named rCAA-8, exhibited a strong binding affinity for the repetitive domain of CA125, enabling differentiation between CA125-expressing cells (OVCAR-3) and CA125-negative cells (SKOV-3) through fluorescence imaging. In comparison to conventional enzyme-linked immunosorbent assays (ELISAs) targeting CA125, the aptamer-based CA125 assay performed on the 3DN-CNTs demonstrated enhanced sensitivity and a wider dynamic range based on the concentration of CA125. This improvement was attributed to the aptamer’s high specificity for the target and the high density of target molecules on the biochip surface. Consequently, the selected aptamer and its biochip applications have promising potential for facilitating the sensitive detection of CA125 in clinical settings
[8].
Tripathi et al. conducted a study in which they developed a new DNA aptamer using a Membrane-SELEX approach. Among the aptamers generated, Apt 2.26 showed the highest affinity and specificity for CA125. To characterize its binding properties, membrane-based assessment was employed to observe the binding of Apt 2.26 to CA125. In vitro methods including DOT ELASA, NALFA, and DPV were utilized to investigate the binding interaction between Apt 2.26 and CA125, demonstrating the diagnostic potential of this screened aptamer. Furthermore, the stability of Apt 2.26 in human serum and high salt concentrations confirmed its robust nature, making it suitable for use with complex sample matrices
[35].
3.3. Human Epididymis Protein 4 (HE4)
Human epididymis protein 4 (HE4), initially identified in the human epididymis, is a 25 kDa serine protease inhibitor that is expressed in the reproductive and respiratory tracts. This protein exhibits interactions with various other proteins, including MUC16 (CA125) and WFDC members like SPINT4 (serine peptidase inhibitor, Kunitz type 4). HE4 is secreted into the bloodstream as a glycoprotein and is found at elevated levels in patients with serous and endometrioid EOC. Notably, HE4 is up regulated in OC, and its elevated levels have been observed in serous, endometrioid, and clear-cell ovarian tumors. Furthermore, HE4 has shown increased expression in patients with mucinous ovarian tumors, which distinguishes it from CA125
[36][37][38][39][40]. In contrast to CA125, HE4 offers several advantages as a biomarker. HE4 serum levels do not increase during pregnancy, menstruation, or in benign gynecological conditions. Additionally, HE4 is elevated at an earlier stage in the progression of the disease
[36][41]. Nevertheless, the concentration of HE4 is substantially influenced by age
[38].
Eaton et al. conducted a study to investigate aptamers that specifically target HE4. The research employed capillary electrophoresis and encompassed five rounds of selection using a single-stranded DNA (ssDNA) library with a 25-nucleotide random region. The ssDNA library was exposed to recombinant glutathione-S-transferase (GST)-HE4 protein. To eliminate aptamers binding to the protein tag, two rounds of counter-selection against GST were performed. Following PCR amplification, the ssDNA oligonucleotides were regenerated using streptavidin columns. Each round of selection was sequenced, and the data were analyzed using a bioinformatics data pipeline called the Rebecca Whelan Python Enrichment. Candidate aptamers were selected for further characterization based on high-fold enrichment and cluster abundance. Notably, aptamers A1, A3, and B10 exhibited affinity towards HE4
[9]. Unexpectedly, the A1 aptamer designed to bind with HE4 has been employed as a recognition probe in aptasensors specifically targeting CA125
[42][43]. In a recent advancement, the A1 aptamer sequence was utilized to create an aptasensor that utilizes up-conversion luminescence resonance energy transfer (LRET) for the detection of HE4
[44].
Hanžek et al. successfully identified novel DNA aptamers, namely AHE1 and AHE3, which demonstrate a high binding affinity to the human HE4 protein present in urine samples. These anti-HE4 aptamers were specifically selected through the implementation of Hi-Fi SELEX, a selection technique that relies on digital droplet PCR partitioning and sensitive amplification methods to isolate rare aptamer sequences. By utilizing this approach, the researchers were able to isolate aptamers that effectively target the HE4 protein within the complex urine environment. Given that HE4 levels exhibit elevation in the urine of patients, the aptamers underwent further characterization specifically in urine samples. The results revealed that AHE1 and AHE3 exhibited a favorable affinity towards the target HE4, as evidenced by a dissociation constant measured within the nanomolar range. These compelling findings strongly indicate the potential of AHE1 and AHE3 as promising diagnostic probes, which could be further harnessed in the development of advanced diagnostic tests and biosensors intended for the reliable detection of OC
[36].
3.4. Epithelial Cell Adhesion Molecules (EpCAM)
Epithelial cell adhesion molecules (EpCAM or CD326) are glycosylated membrane proteins that belong to the small GA733 protein family and are characterized as type I transmembrane glycoproteins. They have been found to be overexpressed in more than 70% of OC cases, and their expression levels are closely linked to the presence of malignant ascites, chemoresistance, and reduced survival rates among OC patients
[45][46][47]. While EpCAM is typically expressed in normal epithelial tissues, its expression within the peritoneal cavity seems to be specific to tumors, as mesothelial cells in the abdominal cavity do not exhibit EpCAM expression. EpCAM plays a pivotal role in driving cancer progression by regulating various cellular attributes, including proliferation, stemness, mobility, and resistance to chemotherapy and radiotherapy. Additionally, the aberrant expression of EpCAM can influence the immune microenvironment, leading to modulations in immune responses
[46][48][49]. It is important to note that the functions of EpCAM are highly reliant on the specific context and exhibit dynamic characteristics. The downregulation of EpCAM has been associated with the inhibition of cell-cell adhesion and the process of epithelial-mesenchymal transition (EMT), suggesting its involvement in the progression of OC
[45][50]. Recent studies have linked EpCAM to oncogenic features, including the enhanced transcription of c-Myc and cyclin A/E. Furthermore, cells expressing EpCAM are known to possess tumor-initiating potential, making EpCAM a significant marker for OC stem cells. These findings provide compelling evidence that EpCAM represents a highly promising and suitable therapeutic target for OC
[45][46][47][48][49][50][51].
Zheng et al. developed a bispecific aptamer designed to simultaneously target EpCAM and CD44, to inhibit cell growth and induce apoptosis in intraperitoneal OC cells. To achieve this, they utilized a double-stranded RNA adaptor to link two aptamers that specifically bind to EpCAM and CD44. This novel approach not only extended the half-life of the aptamers in circulation but also reduced renal filtration compared to using a single aptamer. Through cell-based studies and animal trials, the researchers observed promising results, indicating that the fused aptamer effectively suppressed OC cell growth and inhibited intraperitoneal tumor progression to a greater extent than using CD44 and EpCAM aptamers individually or in combination
[45].
3.5. CD44
CD44, also known as HCAM, Hermes antigen, or lymphocyte homing receptor, is a cell surface glycoprotein that plays a crucial role in mediating cellular responses to the surrounding microenvironment. It is involved in diverse intracellular processes including cell proliferation, survival, motility, and differentiation. Aberrant expression of CD44 has been observed in various epithelial malignancies, including head and neck, colon, and endometrial cancers as well as OC
[52][53][54]. Extensive research has focused on investigating the correlation between CD44 expression or its isoforms and the survival of OC patients
[54]. CD44 is found to be expressed in a majority of EOC tumors, and higher levels of CD44 are associated with advanced disease stages. Activation of the PI3K/Akt and MAPK pathways, facilitated by CD44, may inhibit apoptosis and promote invasion and metastasis of cancer cells. Moreover, CD44 levels serve as effective markers for diagnosing OC and predicting clinical outcomes. Importantly, the upregulation of CD44 in OC is strongly linked to the occurrence of metastasis and disease relapse
[52][53][54][55][56].
The CD44-EpCAM aptamer, developed by Zheng et al., represents a noteworthy CD44-targeting aptamer with the ability to simultaneously bind to EpCAM. This bispecific aptamer demonstrated notable superiority in suppressing the growth of intraperitoneal tumors compared to single aptamers. The augmented efficacy of the bispecific CD44-EpCAM aptamer is primarily attributed to its prolonged circulation time in comparison to the individual aptamers. Importantly, the researchers also determined that the bispecific CD44-EpCAM aptamer exhibited an excellent safety profile as it displayed no signs of toxicity to the host. Furthermore, it was found to be incapable of eliciting innate immunogenic responses, indicating its potential suitability for therapeutic applications without triggering unwanted immune reactions
[45].
3.6. CD5 Antigen-Like Precursor (CD5L)
CD5 antigen-like precursor (CD5L), also known as apoptosis inhibitor of macrophages (AIM), is a 40 kDa soluble glycoprotein that belongs to the scavenger receptor cysteine-rich (SRCR) superfamily. This versatile protein regulates macrophage activity in a broad range of contexts, contributing to the pathogenesis of various infectious and inflammatory conditions. Furthermore, CD5L is implicated in diverse cellular processes, including atherosclerosis and cancer. Additional roles of CD5L have been discovered since, but those related specifically to endothelial cells and angiogenesis remain unknown
[57][58].
LaFargue et al. presented findings that provide evidence for the involvement of CD5L in the development of adaptive resistance to bevacizumab, an antiangiogenic therapy. They demonstrated that neutralizing CD5L using either an antibody or an aptamer effectively inhibited the adaptive resistance to antiangiogenic treatment. The S76.T RNA-aptamer, developed by the researchers, was found to reduce the expression of pAKT and significantly inhibit tube formation and cell migration in AVA-resistant RF24 endothelial cells. To assess the impact of S76.T on adaptive resistance in vivo, SKOV3ip1 OC cells were injected into the peritoneal cavity of nude mice. Mice treated with S76.T aptamer alone exhibited a reduction in tumor burden, while mice treated with a combination of the aptamer and B20—an anti-VEGF (vascular endothelial growth factor) antibody—showed even greater reductions in tumor weight, fewer tumor nodules, as well as lower microvessel density and proliferation compared to mice treated with a control scramble IgG. These collective findings highlight CD5L as a significant protein involved in the adaptive response to anti-VEGF treatment. Consequently, targeting CD5L could be a beneficial strategy for patients undergoing treatment with antiangiogenic drugs
[58].
3.7. Vimentin
Vimentin, an extracellular matrix protein belonging to the intermediate filament protein family, has garnered significant attention in cancer research. Numerous studies have demonstrated a direct association between elevated vimentin expression and reduced survival rates across various cancer types, including colorectal, cervical, breast, gastric, and non-small-cell lung cancers
[59][60][61]. The presence of vimentin filaments within cancer cells serves as a protective mechanism during migration and traversal through narrow spaces, as they establish a viscoelastic framework that can withstand mechanical stresses. Furthermore, vimentin plays a crucial role in maintaining the structural integrity of organelles, particularly the nucleus, during the process of EMT and cancer progression
[62]. Notably, vimentin has been found to safeguard cancer cells against internal stress induced by misfolded proteins by directly binding to stress granules and aggresomes, facilitating their subsequent degradation
[63]. Moreover, vimentin overexpression has been linked to an augmented metastatic capacity attributed to the induction of EMT in ovarian tumor cells
[59]. Apart from its structural functions, vimentin also participates in various signaling pathways associated with cell adhesion, migration, and apoptosis. However, limited research has explored the specific relationship between vimentin expression and ovarian tumor prognosis
[60][62][63][64]. Szubert et al. demonstrated that increased vimentin expression in ovarian tumor cells was correlated with a prolonged overall survival rate
[65].
Costello et al. have discovered two distinct truncated aptamer motifs, namely V3M2 and V5M2, which exhibit effective binding to the extracellular matrix protein vimentin in OC cells and human ovarian tumor tissue. By truncating specific regions within the stem-loop structure, the binding affinity of the aptamers to the vimentin protein was significantly enhanced compared to that of the full-length aptamer sequences V3 and V5
[59].
3.8. Nucleolin
Nucleolin also referred to as C23, is a prominent protein abundantly present in the nucleolus, constituting approximately 10% of the protein content within this subnuclear compartment
[66]. Evidence has established the involvement of nucleolin in crucial cellular processes, including nucleolar chromatin remodeling, pre-RNA maturation, rDNA transcription, and ribosome assembly
[67][68]. In normal cells, nucleolin, characterized by its RNA recognition motifs, predominantly resides within the nucleolus. However, in cancer cells, a remarkable upregulation of nucleolin has been observed on the cell surface
[69]. This abnormal overexpression of nucleolin is closely associated with adverse patient outcomes, as this protein exerts oncogenic effects by promoting carcinogenesis, cellular proliferation, metastasis, and angiogenesis. Notably, nucleolin undergoes extensive phosphorylation, which is tightly regulated throughout the cell cycle. Although no mutations or splicing variants of nucleolin have been linked to malignancy, dysregulated accumulation of nucleolin messenger RNA (mRNA) and/or protein has been reported in various cancer types
[66][67][68].
Ruan et al. successfully assembled six AS1411 aptamers onto two hexangular DNA tiles, resulting in the creation of two distinct aptamers with efficient and divergent structures: HA-6AS, characterized by a three-dimensional tubular shape, and ST-6AS, possessing a two-dimensional six-star configuration. Their investigation revealed that both HA-6AS and ST-6AS exhibited superior capabilities in loading doxorubicin (DOX) compared to DNA tetrahedron structures. Moreover, these aptamers effectively delivered DOX into OC cells, enabling its evasion from lysosomes and subsequent localization within the cell nucleus. HA-6AS-DOX and ST-6AS-DOX demonstrated rapid cytotoxicity specifically against OC cells while exhibiting low levels of cytotoxicity towards normal ovarian epithelial cells. Additionally, the study revealed contrasting internalization patterns between HA-6AS and ST-6AS; HA-6AS displayed a more active targeting mechanism, whereas ST-6AS exhibited a predominantly passive targeting mechanism
[70].
3.9. Stress-Induced Phosphoprotein 1 (STIP1)
STIP1, also referred to as Hsp70/Hsp90-organizing protein (HOP), encompasses three tetratricopeptide repeat domains that enable interactions with heat shock proteins (Hsps), leading to the formation of complexes involved in various biological processes
[71]. These processes encompass RNA splicing, transcription, protein folding, signal transduction, and cell cycle regulation. STIP1 serves as a mediator for protein transfer between Hsp70 and Hsp90, thus facilitating the formation of the Hsp70/Hsp90 complex. This complex plays a critical role in the folding and maturation of transcription factors, hormone receptors, and protein kinases. Hsps are implicated in the initiation, facilitation, and progression of malignant conditions
[72][73][74]. Reports have indicated the overexpression of STIP1 in ovarian, breast, renal, gastric, and colorectal cancers and oral squamous cell carcinoma
[73][74]. In human OC, STIP1 has been identified as a released biomarker. In the context of human OC cells, secreted STIP1 has been found to enhance tumor cell proliferation by binding to ALK2 (activin A receptor, type II-like kinase 2) and activating the SMAD-ID3 signaling pathways. Notably, clinical investigations have revealed that elevated expression of the STIP1 protein is associated with unfavorable prognostic outcomes in cases of OC
[72].
Van Simaeys et al. presented a novel approach for the straightforward identification of the target protein of aptamer 1, which was found to be STIP1. They further revealed the involvement of STIP1 in the invasiveness of the TOV-21G OC cell line. Building upon the identification of STIP1 as a promising biomarker for OC through their methodology, the researchers subsequently explored the therapeutic potential of aptamer TOV6. Remarkably, the aptamer exhibited the capability to inhibit cellular invasion, thus highlighting its potential as a valuable tool for therapeutic intervention
[13].
Chen et al. presented a novel approach for the early diagnosis of OC, involving the respective and simultaneous detection of CA125 and STIP1. They utilized aptamer-based fluorescent and RLS (resonance light scattering) sensors, which provided strong evidence for the presence of OC in its early stages. The findings demonstrated that these sensors exhibited excellent sensitivity and specificity in detecting the tumor markers CA125 and STIP1, thereby offering a promising diagnostic tool for OC
[75].
3.10. Heat Shock Proteins Hsp70 (HSP70s)
The 70 kDa heat shock proteins (HSP70s) constitute a highly conserved and inducible group of heat shock proteins. Their primary role lies in acting as molecular chaperones and participating in various cellular processes that involve protein folding and remodeling
[76][77][78]. HSP70s are frequently observed to be overexpressed and can serve as prognostic markers in numerous cancer types
[78]. Moreover, they play critical roles in the molecular processes underpinning cancer hallmarks, as well as influencing the growth and survival of cancer cells. Interestingly, the impact of HSP70s on cancer cells extends beyond their chaperone activities; they also exert significant regulatory effects on cancer cell signaling. Consequently, HSP70′s high expression in cancer cells is often associated with unfavorable outcomes, including poor prognosis, disease progression, recurrence, and resistance to treatment
[76][77][78][79][80].
Rérole et al. have discovered a range of peptide aptamers capable of binding to HSP70. In the context of human tumor cells, two of these peptide aptamers, namely A8 and A17, exhibited binding affinity to distinct regions of HSP70—the peptide-binding and ATP-binding domains, respectively. Remarkably, these aptamers effectively impeded the chaperone activity of HSP70, thereby heightening the susceptibility of cells to apoptosis induced by anticancer drugs. Significantly, P17, a 13-amino acid peptide derived from the variable region of A17, exhibited persistent specific inhibitory characteristics targeting HSP70. Interestingly, in vivo investigations employing both local and systemic administration of P17 demonstrated substantial regression of subcutaneous tumors. Notably, this therapeutic outcome displayed a noteworthy association with a pronounced influx of macrophages and T lymphocytes into the TME
[77].
3.11. Mucin 1 (MUC1)
MUC1, also known as episialin or EMA, is a prominent member of the mucin family and functions as a transmembrane glycoprotein. Structurally, it forms a type-I transmembrane heterodimer consisting of two subunits that are non-covalently bound together
[81][82]. In normal physiological conditions, MUC1 is primarily expressed on the apical surface of secretory epithelia, including those found in the mammary gland, gastrointestinal tract, respiratory system, urinary system, and reproductive organs
[83]. However, upregulated expression of MUC1 contributes to the advancement of tumors through its influence on multiple signaling pathways as well as its regulation of tumor cell proliferation and epithelial-mesenchymal transition
[84][85][86]. In the context of cancer progression and metastasis, the expression of MUC1 is characterized by elevated levels, altered glycosylation patterns, and abnormal distribution on the cell surface
[87][88]. Notably, the MUC1 oncogene is frequently overexpressed in various epithelial adenocarcinomas such as lung, liver, pancreatic, and breast cancer and OC
[84][89].
Ferreira et al. have successfully engineered a collection of DNA aptamers capable of binding to MUC1 peptides with exceptional affinity and selectivity. Among the assortment of aptamers generated during their investigation, S2.2 exhibited the most favorable K
d, implying a superior binding affinity towards the MUC1 target. Additionally, the researchers conducted a thorough evaluation of the aptamers’ diagnostic assay potential, thereby highlighting their prospective utility in clinical diagnostic applications
[7]. In their subsequent investigation, the research team devised aptamer 5TR1, which selectively targets the five consecutive repeats located within the variable tandem repeat region of MUC1. Subsequently, this aptamer was employed in the creation of a hybrid sandwich ELISA in conjunction with an antibody. This innovative approach facilitated the identification and quantification of MUC1 in buffered solutions. Furthermore, recovery studies conducted under buffer conditions yielded averaged recoveries ranging from 98.2% to 101.7% for all spiked samples, thus substantiating the aptamer’s viability as a receptor in microtiter-based assays. Notably, the developed aptamer exhibited comparable binding characteristics to the corresponding antibody and even competed with the antibody for binding
[90].
3.12. Human Epidermal Growth Factor Receptor 2 (HER2)
Human epidermal growth factor receptor 2 (HER2), also known as Neu or ErbB2, belongs to the family of epidermal growth factor receptors (EGFRs) that are receptor tyrosine kinases. EGFRs play crucial roles in regulating cell proliferation and differentiation during both embryonic development and adult tissue homeostasis
[91]. In numerous cancer types, HER2 exhibits overexpression and is associated with unfavorable prognosis, suboptimal treatment outcomes, and reduced survival rates
[92][93][94]. Correspondingly, overexpression of HER2 in human OC cell lines has been linked to enhanced DNA synthesis, accelerated cell growth, improved efficiency in soft agar cloning, and increased tumorigenicity in xenograft models using nude mice
[91][95]. Various HER2-targeted therapies have been developed to treat HER2-overexpressing tumors, effectively downregulating HER2 expression. Consequently, profiling HER2 expression is of significant importance in cancer prognosis, patient stratification, and monitoring therapy response
[91][92][93].
Varmira et al. developed a modified RNA aptamer conjugated with hynic and labeled with 99m Tc, aiming to create a radiopharmaceutical suitable for diagnostic imaging of OC cells (SKOV-3) exhibiting high expression of HER2. The complex demonstrated stability in normal saline and serum while specifically targeting the HER2 receptor on cancer cells. Upon injection of the
99mTc-hynic-RNA aptamer, rapid clearance from the bloodstream was observed, with predominant excretion occurring through the hepatobiliary system. However, despite the in vitro specificity of the radioconjugated aptamer in binding to the HER2 receptor on cells, no significant tumor-to-blood or tumor-to-muscle ratios were observed. This lack of specificity binding may be attributed to the in vivo biodistribution of the
99mTc-hynic-RNA aptamer. Based on these findings, the team concluded that modifying the radiometal chelator and the structure of the RNA aptamer could potentially improve tissue uptake and enhance the pharmacokinetic properties of the radiolabeled aptamer for improved tumor targeting
[92].
Zhu et al. employed a combinatorial screening approach, both in vitro and in vivo, to identify DNA aptamers targeting HER2 receptors. These aptamers, named Heraptamers, were subsequently labeled with
18F to enable their utilization in positron emission tomography (PET) imaging of HER2 in OC. Initially, the Heraptamers were selected and validated through in vitro assessments involving the HER2 extracellular domain (ECD) and HER2-positive SKOV-3 OC cells. Subsequently, the aptamers were modified with an alkyne, radiolabeled with
18F using azide-functionalized precursors through click chemistry, and evaluated in SKOV3-tumor-bearing mice using PET imaging. Notably, two aptamers, Heraptamer1 and Heraptamer2, exhibited rapid and significant accumulation in tumors within a short timeframe of just 1 h. In contrast, these aptamers demonstrated minimal uptake in control tumors, underscoring their specific targeting capability
[93].
3.13. Programmed Death-Ligand 1 (PD-L1)
Programmed death-ligand 1 (PD-L1) is a transmembrane protein that serves as a major ligand for PD-1. It is characterized by its overexpression in various cancer cells and is recognized as a potential biomarker in cancers exhibiting elevated PD-L1 levels. PD-L1 plays a critical role in modulating the host’s antitumor immune response, enabling tumor cells to evade immune surveillance and promoting metastasis
[11][96][97]. Despite the significance of PD-L1 expression as a prognostic indicator, the immunological pathway associated with high PD-L1 expression in OC remains understudied, and the molecular mechanisms involving immune cells and tumor cells remain elusive
[97][98].
Yazdian-Robati et al. have presented novel DNA aptamers that exhibit highly specific and strong binding affinity towards PD-L1. The researchers evaluated the affinity and specificity of two screened aptamers, namely Apt5 and Apt33. Furthermore, they examined the internalization of Apt5 for potential applications in targeted delivery and imaging. Aptamer Apt5 demonstrated exceptional affinity and selectivity for PD-L1 and was observed to be internalized into OC cells. Additionally, the results indicated that Apt5 successfully detected cancer cells, thus highlighting its potential for early-stage cancer cell detection
[11].
3.14. Kisspeptin-1 (KiSS-1)
Kisspeptin, a polypeptide, and its corresponding encoding gene KiSS-1 were initially identified as an inhibitor of human metastasis and as a gene associated with malignant melanoma
[99]. Intriguingly, recent research has indicated significant alterations in the expression of kisspeptin and its receptor in women affected by various cancer types, including lung cancer, OC, breast cancer, and endometrial cancer. In recent years, extensive investigations have explored the roles of KiSS-1 and KiSS-1R in diverse malignant tumors, highlighting their potential as prognostic or therapeutic markers in cancers
[99][100]. The molecular mechanisms underlying the regulation of cancer cell proliferation and metastasis by the kisspeptin/KiSS-1R system involve intricate endocrine, paracrine, and autocrine actions. Additionally, this system may play a role in modulating tumor tissue angiogenesis. Consistent with these findings, a recent study demonstrated higher preoperative expression of KiSS-1 in ovarian tumor tissue compared to non-tumor tissue. Moreover, the presence of metastasis and tumor size exhibited a negative correlation with preoperative KiSS-1 expression
[99][101][102][103].
Singh et al. focused on the utilization of KiSS-1 as a target for novel and specific Kisspeptin aptamers. The researchers concluded that for diagnosis purposes, it is likely that a combination of kisspeptin levels with established tumor markers would be required, as a singular measurement of kisspeptin may be insufficient. Additionally, they suggested that the development of specific aptamers against the KiSS1R holds potential and could aid in excluding biological interactions with other receptors, such as NPFF1 and NPFF2, which also respond to kisspeptin. The use of highly selective aptamers with a strong affinity for kisspeptin would enable more precise targeting of this molecular target within cancer-related pathophysiological mechanisms. This approach would minimize the risk of nonspecific blockade of other chemical mediators or interference with different signaling pathways involving kisspeptin, thereby reducing the likelihood of adverse effects in patients. Consequently, the application of such specific aptamers in therapeutic strategies for OC holds considerable advantages in the clinical management of these conditions, emphasizing the need for further research in this area
[104].
3.15. Ephrin-A2 (EphA2)
The erythropoietin-producing hepatocellular receptors (EPHs) represent the largest subfamily of receptor tyrosine kinases, engaging with membrane-bound proteins known as ephrins
[105]. EPHs and their ligands are widely expressed, particularly during early development, across various cell types, participating in several physiological functions critical for embryonic development, including the regulation of processes such as cell migration and adhesion
[106][107][108]. In contrast to other EPH kinases, EphA2, a transmembrane protein with a molecular weight of 130 kDa, predominantly resides in adult human epithelial cells. Its expression is typically low
[106][109]. The EphA2 receptor mediates signaling pathways that negatively modulate epithelial cell growth, suppress cell migration, and induce EphA2 internalization and degradation
[110][111]. However, malignant cells often exhibit weakened cell-cell contacts, preventing the association of the EphA2 receptor with ephrin, and leading to upregulated EphA2 receptor expression. High levels of EphA2 expression have been reported in various solid tumors, including ovarian, prostate, pancreatic, lung, esophageal, colorectal, and breast cancers as well as bone sarcomas, melanoma, and glioblastoma
[109]. Furthermore, numerous studies have demonstrated that the overexpression of the EphA2 receptor plays a pivotal role in promoting the aggressiveness and metastatic potential of many of these cancers
[112]. Moreover, EphA2 is closely associated with key regulators of angiogenesis
[113]. Hence, EphA2 represents a critically important therapeutic target, and agents that promote EphA2 activation hold significant potential for suppressing cancer cell malignancy.
Santana-Viera et al. identified a 20-fluoro-modified pyrimidine RNA aptamer named ATOP, which specifically targets EphA2. The ATOP EphA2 aptamer was discovered through the use of an aptamer clustering algorithm, which aimed to identify a common sequence-structure motif derived from two parallel selection processes: protein SELEX 48 involving hEphA2 and cell-internalization SELEX 49 involving MDA231 cells expressing EphA2. When applied to tumor cell lines expressing EphA2, the ATOP EphA2 aptamer effectively reduced tumor cell migration and clonogenicity. In an in vivo mouse model of Ewing sarcoma metastasis, the ATOP EphA2 aptamer exhibited the ability to decrease primary tumor growth and significantly reduce the occurrence of lung metastases. Consequently, the EphA2 ATOP aptamer emerges as a promising candidate for the development of next-generation targeted therapies, offering safer and more effective treatment options for EphA2-overexpressing tumors
[109].
4. Aptamers Enhancing Immunity in OC
Immunotherapeutics represent a revolution in medicine. However, their clinical application is limited in OCs due to the immunosuppressive nature of the TME. Therefore, the identification of potential targets to overcome immunological tolerance and enhance the immune response is one of the challenges facing medicine. Currently, immunotherapy relies on drugs that target immune checkpoints. These approaches can be used in monotherapy as well as in combination therapy, which may include antiangiogenic agents, PARP inhibitors, cisplatin, and more. One of the innovative approaches to enhancing the immune response involves the use of aptamers
[114]. Compared to antibodies, aptamers are more stable, can be modified, and can be used for targeted drug delivery
[6].
Understanding the TME of OC cells allows for the development of therapies aimed at stimulating the immune system to fight cancer. Immunotherapeutic strategies are currently one of the primary research goals of scientists. The latest scientific reports indicate that myeloid-derived suppressor cells (MDSCs) participate in immunosuppression, which may be correlated with high expression of HSP70. HSP70 is a heat shock protein that participates not only in the immune response but also in regulating apoptosis, necrosis, and angiogenesis. A comparison of urine samples from healthy donors and cancer patients revealed a higher concentration of HSP70 in cancer patients, including OC patients. Consequently, Gobbo et al. developed an aptamer, A8, which inhibits the interaction between HSP70 and TLR2, thereby blocking the activation of MDSCs. It has been demonstrated that aptamer A8 leads to the inhibition of STAT3 phosphorylation and the secretion of IL-6 in a medium containing tumor-derived exosomes. In vivo studies showed that a combination of 5-Fluorouracil and cisplatin may decrease the activation of MDSCs
[115]. These experiments have shown novel perspectives on the use of aptamers in the immunotherapy of OC.
T cells assume a crucial role in immune responses. Nevertheless, within the TME, they undergo differentiation, adopting an exhausted phenotype characterized by heightened expression of immune inhibitory checkpoints, including TIM-3. Significantly, TIM-3 assumes an immunosuppressive role in the context of OC. It hinders the intracellular transport of nucleic acids into ovarian tumor cells’ cysts, consequently diminishing the therapeutic efficacy of chemotherapy
[116]. This elevation in TIM-3 expression holds notable significance in the pathogenesis of OC
[117]. In 2017, Gefen et al. designed a TIM-3 aptamer to enhance T-cell cytotoxic activity in a mouse model injected with colon carcinoma. In vitro studies demonstrated the highest increase in the proliferation of CD8+ T cells, which was associated with an upregulation of cytotoxic cytokines, including IFN-γ and TNF-α. Moreover, in vivo studies revealed that mice treated with the aptamer targeting TIM-3 experienced a more significant reduction in tumor volume correlated with an increased number of activated T cells compared to mice treated with a TIM-3 antibody. These results indicate that aptamers may offer a promising approach to activating T cells by inhibiting the TIM-3 receptor
[118].
One of the inhibitors of the immune system response is PD-L1, overexpression of which was found in cancer cells. PD-L1 exerts its inhibitory effects by engaging with the PD-1 receptor, resulting in the attenuation of cytotoxic cytokine production and, consequently, a reduction in the efficacy of T-cell activity. As outlined earlier, Yazdian-Robati et al. designed two aptamers, Apt5 and Apt33, that bind to PD-L1, offering the potential for targeted delivery of anticancer drugs and increasing the immune response. Apt5 exhibited the highest affinity for binding to PD-L1 with high specificity. Furthermore, the researchers demonstrated that labeling this aptamer with ATTO647N could be a promising approach for early-stage OC detection. It is important to highlight that this labeling procedure comes with a significant cost. Therefore, further studies are needed to optimize the production cost of the presented detection method
[11].