
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mark Willcox | -- | 2443 | 2023-11-17 02:00:26 | | | |
| 2 | Catherine Yang | Meta information modification | 2443 | 2023-11-17 02:51:47 | | |
Video Upload Options
The polymyxin antibiotics colistin and polymyxin B have been recently revitalized as bactericidal drugs due to the increase in bacterial resistance to many commonly used antibiotics. Polymyxins were originally derived from the bacterium Paenibacillus polymyxa as the products of fermentation in the form of amphipathic lipopeptide molecules. Polymyxins were discovered in the 1940s to be cyclic lipodecapeptide antibiotics and recognized for therapeutic use in the 1950s. Polymyxins contain conserved components that consist of a d-Phe6-l-Leu7 segment, an N-terminal fatty acyl chain separated by cationic residues (l-α-γ-diaminobutyric acid (Dab)), and segments of the polar amino acid threonine (Thr). Polymyxins target the negatively charged outer membrane lipopolysaccharides (LPSs) of Gram-negative bacteria. Mobilized colistin resistance, mcr, genes are mainly associated with bacterial plasmids. These play an important role in the spread of colistin resistance because of their transferability among different strains in different environments. These mcr genes encode phosphoethanolamine-lipid A transferases that mediate the addition of PEA to the lipid A of an LPS at the 1′ and 4′ positions, causing a significant reduction in the overall negative charge on the bacterial outer membrane. This ultimately leads to the loss of binding affinity of an LPS to the cationic polymyxins and therefore resistance to their action.
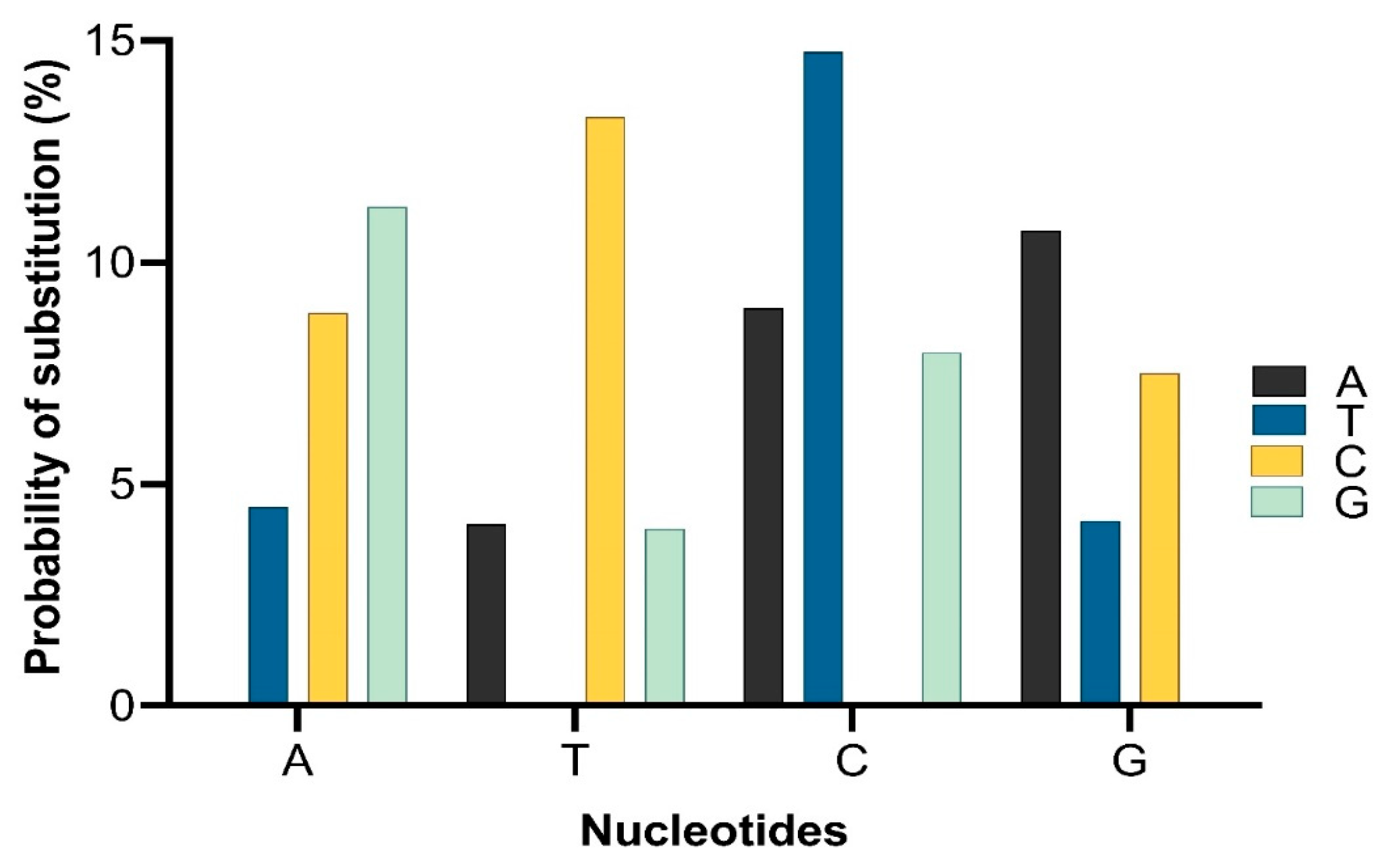
1. Global Dissemination of mcr among Different Bacteria in Different Environments
| mcr Gene Number | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| mcr gene number and source | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| mcr-1 Escherichia coli KU886144.1 | 0.18 | 0.67 | 0.57 | 0.54 | 0.22 | 0.47 | 0.68 | 0.71 | 0.71 | |
| mcr-2 Pseudomonas aeruginosa MW811418.1 | 0.18 | 0.68 | 0.58 | 0.56 | 0.12 | 0.49 | 0.69 | 0.7 | 0.72 | |
| mcr-3 Escherichia coli MW811424.1 | 0.67 | 0.68 | 0.62 | 0.75 | 0.68 | 0.7 | 0.76 | 0.38 | 0.38 | |
| mcr-4 Escherichia coli MW811433.1 | 0.57 | 0.58 | 0.62 | 0.56 | 0.58 | 0.49 | 0.65 | 0.65 | 0.61 | |
| mcr-5.1 Salmonella enterica NG055658.1 | 0.54 | 0.56 | 0.75 | 0.56 | 0.55 | 0.43 | 0.64 | 0.72 | 0.73 | |
| mcr-6.1 Moraxella sp. NG055781.1 | 0.22 | 0.12 | 0.68 | 0.58 | 0.55 | 0.51 | 0.72 | 0.72 | 0.74 | |
| mcr-7 Pseudomonas aeruginosa MW811434.1 | 0.47 | 0.49 | 0.7 | 0.49 | 0.43 | 0.51 | 0.65 | 0.71 | 0.68 | |
| mcr-8 Klebsiella pneumoniae MT815555.1 | 0.68 | 0.69 | 0.76 | 0.65 | 0.64 | 0.72 | 0.65 | 0.69 | 0.72 | |
| mcr-9 Uncultured bacterium MW478857.1 | 0.71 | 0.7 | 0.38 | 0.65 | 0.72 | 0.72 | 0.71 | 0.69 | 0.22 | |
| mcr-10.1 Enterobacter cloacae MN044989.1 | 0.71 | 0.72 | 0.38 | 0.61 | 0.73 | 0.74 | 0.68 | 0.72 | 0.22 | |
| Average evolutionary divergence | 0.53 | 0.52 | 0.62 | 0.59 | 0.61 | 0.54 | 0.57 | 0.69 | 0.61 | 0.61 |
| Standard Deviation | 0.20 | 0.23 | 0.14 | 0.05 | 0.11 | 0.23 | 0.11 | 0.04 | 0.18 | 0.19 |
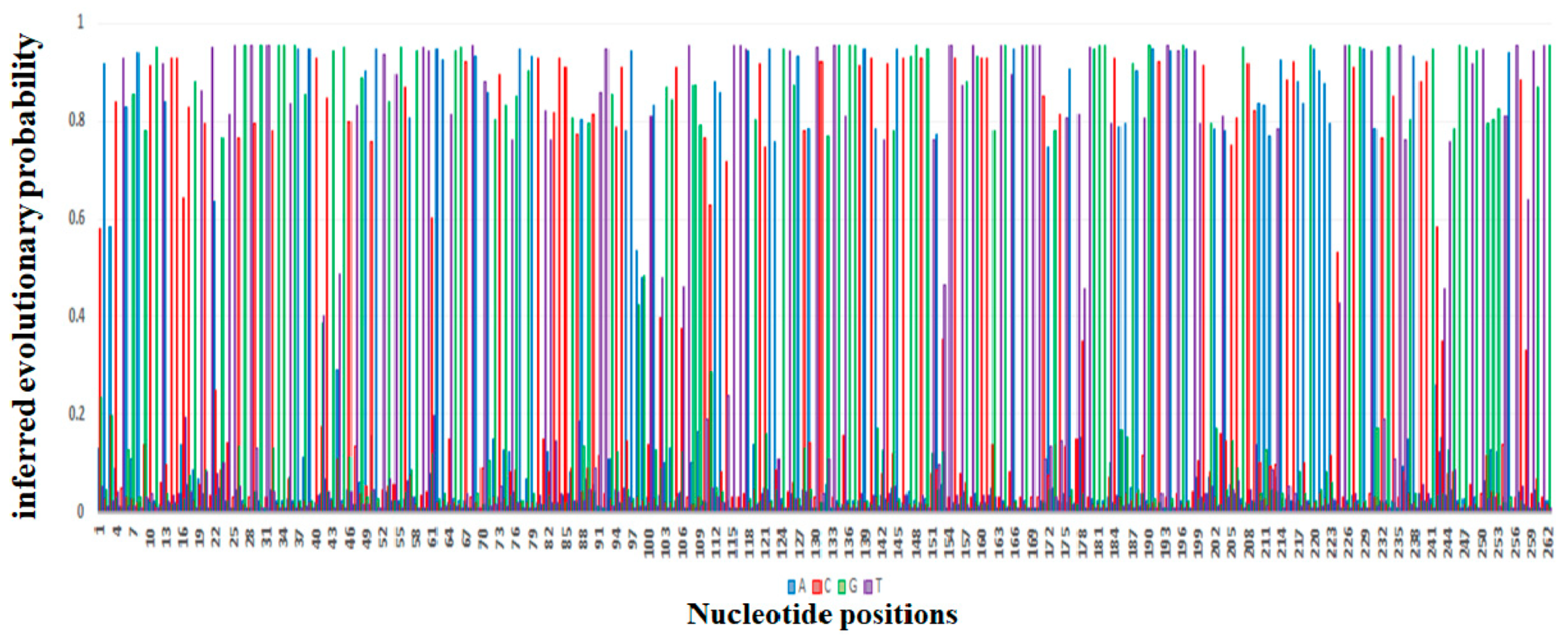
2. Evolution of mcr Gene Variants from mcr-1 to mcr-10
The Processes and Molecular Vehicles Responsible for the Transmission of mcr Variants
| mcr Variants | Insertion Sequences Structure | Transposon | Plasmids | Organism | Host (Isolated from) |
Year of Discovery | References |
|---|---|---|---|---|---|---|---|
| mcr-1 | (ISApl1-mcr-1-pap2-ISApl1 and Tn7511) | Novel transposon Tn7511 | IncI1 plasmid, pMCR-E2899 | E. coli DH5α | Turkey meat | 2022 | [67] |
| mcr-1 | Combination of ISApl1 and IS91 (ISApl1-mcr-1-IS91) | Chromosomal Tn6330 transposon | IncI2 plasmid | E. coli | Community and hospital settings | 2022 | [58] |
| mcr-1 | IS26-mcr-1-PAP2, and ISAPl1-mcr-1-PAP2 and ISEcp1-blaCTX₋M₋₅₅-mcr-1-PAP2 |
--- | IncI2, IncX4, and IncHI2 plasmids | E. coli and Salmonella spp. | Food products, food supply chain, and clinical samples | 2021 | [68][69] |
| mcr-1.1 | IS26-parA-mcr-1.1-pap2 | --- | IncX4-type plasmid | E. coli | Dog feces | 2020 | [56] |
| mcr-1 | I ISApl1-mcr-1-orf ISApl1 | ISApl1 transposon | IncHI2 and IncX4 plasmids | Enterobacteriaceae | Livestock | 2018 | [70] |
| mcr-1 | ISApl1-mcr-1-pap2-ISApl1 | Tn6330 | IncI2 and IncX4 plasmids | Novel Moraxella spp. | Pig | 2018 | [46] |
| mcr-1 | mcr-1-orf, ISApl1-mcr-1-orf and Tn6330 | Novel transposon Tn6330 | IncX4 and IncI2 plasmids | E. coli | Pig farms in China | 2017 | [69] |
| mcr-2 | (ISEc69-mcr-2-ORF-ISEc69 | Tn7052 | IncX4 conjugative plasmid | Moraxella osloensis | --- | 2021 | [71] |
| mcr-2 | ISEc69-mcr-2-ISEc69 | --- | IncX4 plasmid | M. bovoculi | Pigs, pork and chicken meat, and humans | 2017 | [72] |
| mcr- 3.1 | TnAs2-mcr-3.1-dgkA-ISKpn40 | Novel transposon Tn6330 | pCP61-IncFIB plasmid | E. coli | Pigs | 2021 | [73] |
| mcr-3.5 | IS4321R-TnAs2-mcr-3.5-dgkA-IS15 | Novel transposon Tn6330 | IncFIItype plasmid pCP55-IncFII | E. coli | Pigs | 2021 | [73] |
| mcr-3.7 | TnAs2-mcr-3.7-dgkA-IS26 | --- | IncP1 plasmid | E. coli | Dogs | 2020 | [56] |
| mcr-8 | IS903B-ampC-hp-hphp-Giy-T-dgkA-baeS-copR-IS3-mcr-8-Gly-T-IS5 | _ Δ IS66 transposases | IncFIA plasmid | K. pneumoniae | Patients from intensive care | 2022 | [74] |
| mcr-8 | IS903B-ymoA-inhA-mcr-8-copR-baeS-dgkA-ampC | Composite transposon | pABC264-OXA-181 plasmid | K. pneumoniae | Patient with bacteremia | 2022 | [75] |
| mcr-8.2 | ISEcl1-mcr-8.2-orf-ISKpn26 | --- | IncFII/FIA | K. pneumoniae | Patient’s Intestinal sample | 2022 | [76] |
| mcr-9.1 | IS903B-mcr-9.1-wbuC-IS26 | Tn6360 | IncHI2/2A plasmid | E. cloacae complex | Clinical isolates | 2022 | [77] |
| mcr-10 | ISKpn26 is present at upstream of xerC-mcr-10 and an IS26 | Transposon Tn1722 | IncFIA plasmid | Enterobacter roggenkampii | Clinical isolate | 2020 | [78] |
| mcr-10.1 | hsdSMR-ISEc36-mcr-10.1-xerC | --- | IncFIIK plasmids | K. pneumoniae | Clinical isolates | 2022 | [77] |
3. Methods for Detecting Polymyxin Resistance
References
- Shen, Z.; Wang, Y.; Shen, Y.; Shen, J.; Wu, C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 2016, 16, 293.
- Wang, R.; Liu, Y.; Zhang, Q.; Jin, L.; Wang, Q.; Zhang, Y.; Wang, X.; Hu, M.; Li, L.; Qi, J.; et al. The prevalence of colistin resistance in Escherichia coli and Klebsiella pneumoniae isolated from food animals in China: Coexistence of mcr-1 and bla(NDM) with low fitness cost. Int. J. Antimicrob. Agents 2018, 51, 739–744.
- Kuo, S.C.; Huang, W.C.; Wang, H.Y.; Shiau, Y.R.; Cheng, M.F.; Lauderdale, T.L. Colistin resistance gene mcr-1 in Escherichia coli isolates from humans and retail meats, Taiwan. J. Antimicrob. Chemother. 2016, 71, 2327–2329.
- Buess, S.; Nüesch-Inderbinen, M.; Stephan, R.; Zurfluh, K. Assessment of animals as a reservoir for colistin resistance: No MCR-1/MCR-2-producing Enterobacteriaceae detected in Swiss livestock. J. Glob. Antimicrob. Resist. 2017, 8, 33–34.
- Girardello, R.; Piroupo, C.M.; Martins, J., Jr.; Maffucci, M.H.; Cury, A.P.; Franco, M.R.G.; Malta, F.M.; Rocha, N.C.; Pinho, J.R.R.; Rossi, F.; et al. Genomic characterization of mcr-1.1-producing Escherichia coli recovered from human infections in São Paulo, Brazil. Front. Microbiol. 2021, 12, 663414.
- Figueiredo, R.; Card, R.M.; Nunez, J.; Pomba, C.; Mendonça, N.; Anjum, M.F.; Da Silva, G.J. Detection of an mcr-1-encoding plasmid mediating colistin resistance in Salmonella enterica from retail meat in Portugal. J. Antimicrob. Chemother. 2016, 71, 2338–2340.
- Gogry, F.A.; Siddiqui, M.T.; Haq, Q.M.R. Emergence of mcr-1 conferred colistin resistance among bacterial isolates from urban sewage water in India. Environ. Sci. Pollut. Res. Int. 2019, 26, 33715–33717.
- Bilal, H.; Rehman, T.U.; Khan, M.A.; Hameed, F.; Jian, Z.G.; Han, J.; Yang, X. Molecular epidemiology of mcr-1, bla (KPC-2,) and bla (NDM-1) harboring clinically isolated Escherichia coli from Pakistan. Infect. Drug Resist. 2021, 14, 1467–1479.
- Vu Thi Ngoc, B.; Le Viet, T.; Nguyen Thi Tuyet, M.; Nguyen Thi Hong, T.; Nguyen Thi Ngoc, D.; Le Van, D.; Chu Thi, L.; Tran Huy, H.; Penders, J.; Wertheim, H.; et al. Characterization of genetic elements carrying mcr-1 gene in Escherichia coli from the community and hospital settings in Vietnam. Microbiol. Spectr. 2022, 10, e0135621.
- Hadjadj, L.; Baron, S.A.; Olaitan, A.O.; Morand, S.; Rolain, J.M. Co-occurrence of variants of mcr-3 and mcr-8 Genes in a Klebsiella pneumoniae isolate from Laos. Front. Microbiol. 2019, 10, 2720.
- McGann, P.; Snesrud, E.; Maybank, R.; Corey, B.; Ong, A.C.; Clifford, R.; Hinkle, M.; Whitman, T.; Lesho, E.; Schaecher, K.E. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: First report of mcr-1 in the United States. Antimicrob. Agents Chemother. 2016, 60, 4420–4421.
- Cannatelli, A.; Giani, T.; Antonelli, A.; Principe, L.; Luzzaro, F.; Rossolini, G.M. First detection of the mcr-1 colistin resistance gene in Escherichia coli in Italy. Antimicrob. Agents Chemother. 2016, 60, 3257–3258.
- Kawanishi, M.; Abo, H.; Ozawa, M.; Uchiyama, M.; Shirakawa, T.; Suzuki, S.; Shima, A.; Yamashita, A.; Sekizuka, T.; Kato, K.; et al. Prevalence of colistin resistance gene mcr-1 and absence of mcr-2 in Escherichia coli isolated from healthy food-producing animals in Japan. Antimicrob. Agents Chemother. 2017, 61, e02057-16.
- Bhat, A.H. Bacterial zoonoses transmitted by household pets and as reservoirs of antimicrobial resistant bacteria. Microb. Pathog. 2021, 155, 104891.
- Skarżyńska, M.; Zaja, C.M.; Bomba, A.; Bocian, Ł.; Kozdruń, W.; Polak, M.; Wia Cek, J.; Wasyl, D. Antimicrobial resistance glides in the Sky-Free-Living Birds as a reservoir of resistant Escherichia coli with zoonotic potential. Front. Microbiol. 2021, 12, 656223.
- Zurfluh, K.; Nüesch-Inderbinen, M.; Klumpp, J.; Poirel, L.; Nordmann, P.; Stephan, R. Key features of mcr-1-bearing plasmids from Escherichia coli isolated from humans and food. Antimicrob. Resist. Infect. Control 2017, 6, 91.
- Fernandes, M.R.; Sellera, F.P.; Esposito, F.; Sabino, C.P.; Cerdeira, L.; Lincopan, N. Colistin-resistant mcr-1-positive Escherichia coli on public beaches, an infectious threat emerging in recreational waters. Antimicrob. Agents Chemother. 2017, 61, e00234-17.
- Zhao, F.; Feng, Y.; Lü, X.; McNally, A.; Zong, Z. IncP plasmid carrying colistin resistance gene mcr-1 in Klebsiella pneumoniae from hospital sewage. Antimicrob. Agents Chemother. 2017, 61, e02229-16.
- Hembach, N.; Schmid, F.; Alexander, J.; Hiller, C.; Rogall, E.T.; Schwartz, T. Occurrence of the mcr-1 colistin resistance gene and other clinically relevant antibiotic resistance genes in microbial populations at different municipal wastewater treatment plants in Germany. Front. Microbiol. 2017, 8, 1282.
- Sun, P.; Bi, Z.; Nilsson, M.; Zheng, B.; Berglund, B.; Stålsby Lundborg, C.; Börjesson, S.; Li, X.; Chen, B.; Yin, H.; et al. Occurrence of bla(KPC-2), bla(CTX-M), and mcr-1 in Enterobacteriaceae from Well Water in Rural China. Antimicrob. Agents Chemother. 2017, 61, e02569-16.
- Zhang, J.; Wang, J.; Chen, L.; Yassin, A.K.; Kelly, P.; Butaye, P.; Li, J.; Gong, J.; Cattley, R.; Qi, K.; et al. Housefly (Musca domestica) and blow fly (Protophormia terraenovae) as vectors of bacteria carrying colistin resistance genes. Appl. Environ. Microbiol. 2018, 84, e01736-17.
- Bean, D.C.; Wigmore, S.M.; Abdul Momin, M.H.F.; Wareham, D.W. Polymyxin resistant bacteria in Australian poultry. Front. Sustain. Food Syst. 2020, 4, 550318.
- Yoon, E.J.; Hong, J.S.; Yang, J.W.; Lee, K.J.; Lee, H.; Jeong, S.H. Detection of mcr-1 plasmids in Enterobacteriaceae isolates from human specimens: Comparison with those in Escherichia coli isolates from livestock in Korea. Ann. Lab. Med. 2018, 38, 555–562.
- Zeng, K.J.; Doi, Y.; Patil, S.; Huang, X.; Tian, G.B. Emergence of the plasmid-mediated mcr-1 gene in colistin-resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob. Agents Chemother. 2016, 60, 3862–3863.
- Liu, B.T.; Song, F.J.; Zou, M.; Hao, Z.H.; Shan, H. Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and in Escherichia coli from the same animal. Antimicrob. Agents Chemother. 2017, 61, 01444-16.
- Li, X.P.; Fang, L.X.; Jiang, P.; Pan, D.; Xia, J.; Liao, X.P.; Liu, Y.H.; Sun, J. Emergence of the colistin resistance gene mcr-1 in Citrobacter freundii. Int. J. Antimicrob. Agents 2017, 49, 786–787.
- Mendes, A.C.; Novais, Â.; Campos, J.; Rodrigues, C.; Santos, C.; Antunes, P.; Ramos, H.; Peixe, L. mcr-1 in carbapenemase-producing Klebsiella pneumoniae with hospitalized patients, Portugal, 2016–2017. Emerg. Infect. Dis. 2018, 24, 762–766.
- Yi, L.; Wang, J.; Gao, Y.; Liu, Y.; Doi, Y.; Wu, R.; Zeng, Z.; Liang, Z.; Liu, J.H. mcr-1-harboring Salmonella enterica serovar Typhimurium sequence type 34 in pigs, China. Emerg. Infect. Dis. 2017, 23, 291–295.
- Ma, Q.; Huang, Y.; Wang, J.; Xu, X.; Hawkey, J.; Yang, C.; Liang, B.; Hu, X.; Wu, F.; Yang, X.; et al. Multidrug-resistant Shigella sonnei carrying the plasmid-mediated mcr-1 gene in China. Int. J. Antimicrob. Agents 2018, 52, 14–21.
- Luo, J.; Yao, X.; Lv, L.; Doi, Y.; Huang, X.; Huang, S.; Liu, J.H. Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli isolates from retail vegetables in China. Antimicrob. Agents Chemother. 2017, 61, e01139-17.
- He, Z.; Yang, Y.; Li, W.; Ma, X.; Zhang, C.; Zhang, J.; Sun, B.; Ding, T.; Tian, G.B. Comparative genomic analyses of polymyxin-resistant Enterobacteriaceae strains from China. BMC Genom. 2022, 23, 88.
- Ellem, J.A.; Ginn, A.N.; Chen, S.C.; Ferguson, J.; Partridge, S.R.; Iredell, J.R. Locally acquired mcr-1 in Escherichia coli, Australia, 2011 and 2013. Emerg. Infect. Dis. 2017, 23, 1160–1163.
- Bell, J.M.; Lubian, A.F.; Partridge, S.R.; Gottlieb, T.; Iredell, J.; Daley, D.A.; Coombs, G.W. Australian Group on Antimicrobial Resistance (AGAR) Australian Gram-negative Sepsis Outcome Programme (GnSOP) Annual Report 2020. Commun. Dis. Intell. 2022, 46, 1–12.
- Arnott, A.; Wang, Q.; Bachmann, N.; Sadsad, R.; Biswas, C.; Sotomayor, C.; Howard, P.; Rockett, R.; Wiklendt, A.; Iredell, J.R.; et al. Multidrug-resistant Salmonella enterica 4,,12:i:- Sequence Type 34, New South Wales, Australia, 2016–2017. Emerg. Infect. Dis. 2018, 24, 751.
- Ingle, D.J.; Ambrose, R.L.; Baines, S.L.; Duchene, S.; Gonçalves da Silva, A.; Lee, D.Y.J.; Jones, M.; Valcanis, M.; Taiaroa, G.; Ballard, S.A.; et al. Evolutionary dynamics of multidrug resistant Salmonella enterica serovar 4,,12:i:- in Australia. Nat. Commun. 2021, 12, 4786.
- Xiang, R.; Liu, B.H.; Zhang, A.Y.; Lei, C.W.; Ye, X.L.; Yang, Y.X.; Chen, Y.P.; Wang, H.N. Colocation of the polymyxin resistance gene mcr-1 and a variant of mcr-3 on a plasmid in an Escherichia coli isolate from a chicken farm. Antimicrob. Agents Chemother. 2018, 62, e00501-18.
- Belaynehe, K.M.; Shin, S.W.; Park, K.Y.; Jang, J.Y.; Won, H.G.; Yoon, I.J.; Yoo, H.S. Emergence of mcr-1 and mcr-3 variants coding for plasmid-mediated colistin resistance in Escherichia coli isolates from food-producing animals in South Korea. Int. J. Infect. Dis. 2018, 72, 22–24.
- Sun, J.; Fang, L.X.; Wu, Z.; Deng, H.; Yang, R.S.; Li, X.P.; Li, S.M.; Liao, X.P.; Feng, Y.; Liu, Y.H. Genetic analysis of the IncX4 plasmids: Implications for a unique pattern in the mcr-1 acquisition. Sci. Rep. 2017, 7, 424.
- Zhang, J.; Chen, L.; Wang, J.; Yassin, A.K.; Butaye, P.; Kelly, P.; Gong, J.; Guo, W.; Li, J.; Li, M.; et al. Molecular detection of colistin resistance genes (mcr-1, mcr-2 and mcr-3) in nasal/oropharyngeal and anal/cloacal swabs from pigs and poultry. Sci. Rep. 2018, 8, 3705.
- Martins-Sorenson, N.; Snesrud, E.; Xavier, D.E.; Cacci, L.C.; Iavarone, A.T.; McGann, P.; Riley, L.W.; Moreira, B.M. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J. Antimicrob. Chemother. 2020, 75, 60–64.
- Patel, R.; Kumar, S. On estimating evolutionary probabilities of population variants. BMC Evol. Biol. 2019, 19, 133.
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000.
- Tamura, K.; Tao, Q.; Kumar, S. Theoretical Foundation of the RelTime method for estimating divergence times from variable evolutionary rates. Mol. Biol. Evol. 2018, 35, 1770–1782.
- Humphrey, S.; Fillol-Salom, A.; Quiles-Puchalt, N.; Ibarra-Chávez, R.; Haag, A.F.; Chen, J.; Penadés, J.R. Bacterial chromosomal mobility via lateral transduction exceeds that of classical mobile genetic elements. Nat. Commun. 2021, 12, 6509.
- El-Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885.
- Strepis, N.; Voor In ‘t Holt, A.F.; Vos, M.C.; Zandijk, W.H.A.; Heikema, A.P.; Hays, J.P.; Severin, J.A.; Klaassen, C.H.W. Genetic analysis of mcr-1-carrying plasmids from Gram-negative bacteria in a Dutch tertiary care hospital: Evidence for intrapatient and interspecies transmission events. Front. Microbiol. 2021, 12, 727435.
- Goodman, R.N.; Tansirichaiya, S.; Brouwer, M.S.M.; Roberts, A.P. Intracellular transposition of mobile genetic elements associated with the colistin resistance gene mcr-1. Microbiol. Spectr. 2023, 11, e0327822.
- Wang, Q.; Sun, J.; Li, J.; Ding, Y.; Li, X.P.; Lin, J.; Hassan, B.; Feng, Y. Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome 2017, 5, 70.
- Sellera, F.P.; Fernandes, M.R.; Sartori, L.; Carvalho, M.P.; Esposito, F.; Nascimento, C.L.; Dutra, G.H.; Mamizuka, E.M.; Pérez-Chaparro, P.J.; McCulloch, J.A.; et al. Escherichia coli carrying IncX4 plasmid-mediated mcr-1 and blaCTX-M genes in infected migratory Magellanic penguins (Spheniscus magellanicus). J. Antimicrob. Chemother. 2017, 72, 1255–1256.
- Maluta, R.P.; Logue, C.M.; Casas, M.R.; Meng, T.; Guastalli, E.A.; Rojas, T.C.; Montelli, A.C.; Sadatsune, T.; de Carvalho Ramos, M.; Nolan, L.K.; et al. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS ONE 2014, 9, e105016.
- Mshana, S.E.; Imirzalioglu, C.; Hain, T.; Domann, E.; Lyamuya, E.F.; Chakraborty, T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin. Microbiol. Infect. 2011, 17, 1279–1282.
- Guenther, S.; Falgenhauer, L.; Semmler, T.; Imirzalioglu, C.; Chakraborty, T.; Roesler, U.; Roschanski, N. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J. Antimicrob. Chemother. 2017, 72, 1289–1292.
- Wang, Y.; Zhang, R.; Li, J.; Wu, Z.; Yin, W.; Schwarz, S.; Tyrrell, J.M.; Zheng, Y.; Wang, S.; Shen, Z.; et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017, 2, 16260.
- El Garch, F.; Sauget, M.; Hocquet, D.; LeChaudee, D.; Woehrle, F.; Bertrand, X. mcr-1 is borne by highly diverse Escherichia coli isolates since 2004 in food-producing animals in Europe. Clin. Microbiol. Infect. 2017, 23, 51.e51–51.e54.
- Boueroy, P.; Wongsurawat, T.; Jenjaroenpun, P.; Chopjitt, P.; Hatrongjit, R.; Jittapalapong, S.; Kerdsin, A. Plasmidome in mcr-1 harboring carbapenem-resistant Enterobacterales isolates from human in Thailand. Sci. Rep. 2022, 12, 19051.
- Wang, R.; van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1179.
- Matamoros, S.; van Hattem, J.M.; Arcilla, M.S.; Willemse, N.; Melles, D.C.; Penders, J.; Vinh, T.N.; Thi Hoa, N.; Bootsma, M.C.J.; van Genderen, P.J.; et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 2017, 7, 15364.
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168.
- Wang, Q.; Sun, J.; Ding, Y.; Li, X.P.; Liu, Y.H.; Feng, Y. Genomic insights into mcr-1-positive plasmids carried by colistin-resistant Escherichia coli isolates from inpatients. Antimicrob. Agents Chemother. 2017, 61, e00361-17.
- Tegetmeyer, H.E.; Jones, S.C.; Langford, P.R.; Baltes, N. ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet. Microbiol. 2008, 128, 342–353.
- Geurts, A.M.; Hackett, C.S.; Bell, J.B.; Bergemann, T.L.; Collier, L.S.; Carlson, C.M.; Largaespada, D.A.; Hackett, P.B. Structure-based prediction of insertion-site preferences of transposons into chromosomes. Nucleic Acids Res. 2006, 34, 2803–2811.
- Sun, J.; Xu, Y.; Gao, R.; Lin, J.; Wei, W.; Srinivas, S.; Li, D.; Yang, R.S.; Li, X.P.; Liao, X.P.; et al. Deciphering MCR-2 colistin resistance. mBio 2017, 8, e00625-17.
- Xavier, B.B.; Lammens, C.; Ruhal, R.; Kumar-Singh, S.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. EuroSurveill 2016, 21, 30280.
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320.
- Cain, A.K.; Liu, X.; Djordjevic, S.P.; Hall, R.M. Transposons related to Tn1696 in IncHI2 plasmids in multiply antibiotic resistant Salmonella enterica serovar Typhimurium from Australian animals. Microb. Drug Resist. 2010, 16, 197–202.
- Snesrud, E.; McGann, P.; Chandler, M. The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: The vehicle for transferable colistin resistance. mBio 2018, 9, e02381-17.
- Li, W.; Yan, Y.; Chen, J.; Sun, R.; Wang, Y.; Wang, T.; Feng, Z.; Peng, K.; Wang, J.; Chen, S.J. Genomic characterization of conjugative plasmids carrying the mcr-1 gene in foodborne and clinical strains of Salmonella and Escherichia coli. Food Control. 2021, 125, 108032.
- Du, C.; Feng, Y.; Wang, G.; Zhang, Z.; Hu, H.; Yu, Y.; Liu, J.; Qiu, L.; Liu, H.; Guo, Z.; et al. Co-occurrence of the mcr-1.1 and mcr-3.7 genes in a multidrug-resistant Escherichia coli isolate from China. Infect. Drug Resist. 2020, 13, 3649–3655.
- He, Y.Z.; Long, T.F.; He, B.; Li, X.P.; Li, G.; Chen, L.; Liao, X.P.; Liu, Y.H.; Sun, J. ISEc69-mediated mobilization of the colistin resistance gene mcr-2 in Escherichia coli. Front. Microbiol. 2020, 11, 564973.
- Li, R.; Xie, M.; Zhang, J.; Yang, Z.; Liu, L.; Liu, X.; Zheng, Z.; Chan, E.W.; Chen, S. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 2017, 72, 393–401.
- Partridge, S.R. mcr-2 in the IncX4 plasmid pKP37-BE is flanked by directly oriented copies of ISEc69. J. Antimicrob. Chemother. 2017, 72, 1533–1535.
- Li, R.; Du, P.; Zhang, P.; Li, Y.; Yang, X.; Wang, Z.; Wang, J.; Bai, L. Comprehensive genomic investigation of coevolution of mcr genes in Escherichia coli strains via nanopore sequencing. Glob. Chall. 2021, 5, 2000014.
- Bai, S.C.; Li, R.B.; Yang, Y.; Liao, X.P. Sporadic dissemination of mcr-8-ST11 Klebsiella pneumoniae isolates in China. Enferm. Infecc. Microbiol. Clin. 2022, 40, 95–97.
- Ge, H.; Qiao, J.; Xu, H.; Liu, R.; Chen, R.; Li, C.; Hu, X.; Zhou, J.; Guo, X.; Zheng, B. First report of Klebsiella pneumoniae co-producing OXA-181, CTX-M-55, and MCR-8 isolated from the patient with bacteremia. Front. Microbiol. 2022, 13, 1020500.
- Liu, C.; Wu, Y.; Fang, Y.; Sang, Z.; Huang, L.; Dong, N.; Zeng, Y.; Lu, J.; Zhang, R.; Chen, G. Emergence of an ST1326 (CG258) multi-drug resistant Klebsiella pneumoniae co-harboring mcr-8.2, ESBL genes, and the resistance-nodulation-division efflux pump gene cluster tmexCD1-toprJ1 in China. Front. Microbiol. 2022, 13, 800993.
- Jiang, S.; Wang, X.; Yu, H.; Zhang, J.; Wang, J.; Li, J.; Li, X.; Hu, K.; Gong, X.; Gou, X.; et al. Molecular antibiotic resistance mechanisms and co-transmission of the mcr-9 and metallo-β-lactamase genes in carbapenem-resistant Enterobacter cloacae complex. Front. Microbiol. 2022, 13, 1032833.
- Liu, M.C.; Jian, Z.; Liu, W.; Li, J.; Pei, N. One healthaAnalysis of mcr-carrying plasmids and emergence of mcr-10.1 in three species of Klebsiella recovered from humans in China. Microbiol. Spectr. 2022, 10, e0230622.
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516.
- Abdul Momin, M.H.F.; Bean, D.C.; Hendriksen, R.S.; Haenni, M.; Phee, L.M.; Wareham, D.W. CHROMagar COL-APSE: A selective bacterial culture medium for the isolation and differentiation of colistin-resistant Gram-negative pathogens. J. Med. Microbiol. 2017, 66, 1554–1561.
- Przybysz, S.M.; Correa-Martinez, C.; Köck, R.; Becker, K.; Schaumburg, F. SuperPolymyxin™ medium for the screening of colistin-resistant gram-negative bacteria in stool samples. Front. Microbiol. 2018, 9, 2809.
- Bardet, L.; Le Page, S.; Leangapichart, T.; Rolain, J.M. LBJMR medium: A new polyvalent culture medium for isolating and selecting vancomycin and colistin-resistant bacteria. BMC Microbiol. 2017, 17, 220.
- Zhou, M.; Wang, Y.; Liu, C.; Kudinha, T.; Liu, X.; Luo, Y.; Yang, Q.; Sun, H.; Hu, J.; Xu, Y.C. Comparison of five commonly used automated susceptibility testing methods for accuracy in the China Antimicrobial Resistance Surveillance System (CARSS) hospitals. Infect. Drug Resist. 2018, 11, 1347–1358.
- Cordovana, M.; Ambretti, S. Antibiotic susceptibility testing of anaerobic bacteria by broth microdilution method using the MICRONAUT-S Anaerobes MIC plates. Anaerobe 2020, 63, 102217.
- Carretto, E.; Brovarone, F.; Russello, G.; Nardini, P.; El-Bouseary, M.M.; Aboklaish, A.F.; Walsh, T.R.; Tyrrell, J.M. Clinical validation of SensiTest colistin, a broth microdilution-based nethod to evaluate colistin MICs. J. Clin. Microbiol. 2018, 56, e01523-17.
- Poirel, L.; Larpin, Y.; Dobias, J.; Stephan, R.; Decousser, J.W.; Madec, J.Y.; Nordmann, P. Rapid Polymyxin NP test for the detection of polymyxin resistance mediated by the mcr-1/mcr-2 genes. Diagn. Microbiol. Infect. Dis. 2018, 90, 7–10.
- Jouy, E.; Haenni, M.; Le Devendec, L.; Le Roux, A.; Châtre, P.; Madec, J.Y.; Kempf, I. Improvement in routine detection of colistin resistance in E. coli isolated in veterinary diagnostic laboratories. J. Microbiol. Methods 2017, 132, 125–127.
- Coppi, M.; Cannatelli, A.; Antonelli, A.; Baccani, I.; Di Pilato, V.; Sennati, S.; Giani, T.; Rossolini, G.M. A simple phenotypic method for screening of MCR-1-mediated colistin resistance. Clin. Microbiol. Infect. 2018, 24, 201.e201–201.e203.
- Kon, H.; Dalak, M.A.B.; Schwartz, D.; Carmeli, Y.; Lellouche, J. Evaluation of the MICRONAUT MIC-strip colistin assay for colistin susceptibility testing of carbapenem-resistant Acinetobacter baumannii and Enterobacterales. Diagn. Microbiol. Infect. Dis. 2021, 100, 115391.
- Bardet, L.; Okdah, L.; Le Page, S.; Baron, S.A.; Rolain, J.M. Comparative evaluation of the UMIC Colistine kit to assess MIC of colistin of gram-negative rods. BMC Microbiol. 2019, 19, 60.
- Sękowska, A.; Bogiel, T. The Evaluation of Eazyplex® SuperBug CRE assay usefulness for the detection of ESBLs and carbapenemases genes directly from urine samples and positive blood cultures. Antibiotics 2022, 11, 138.
- Chabou, S.; Leangapichart, T.; Okdah, L.; Le Page, S.; Hadjadj, L.; Rolain, J.M. Real-time quantitative PCR assay with Taqman® probe for rapid detection of MCR-1 plasmid-mediated colistin resistance. New Microbes New Infect. 2016, 13, 71–74.
- Zhong, L.L.; Zhou, Q.; Tan, C.Y.; Roberts, A.P.; El-Sayed Ahmed, M.A.E.; Chen, G.; Dai, M.; Yang, F.; Xia, Y.; Liao, K.; et al. Multiplex loop-mediated isothermal amplification (multi-LAMP) assay for rapid detection of mcr-1 to mcr-5 in colistin-resistant bacteria. Infect. Drug Resist. 2019, 12, 1877–1887.
- Borowiak, M.; Baumann, B.; Fischer, J.; Thomas, K.; Deneke, C.; Hammerl, J.A.; Szabo, I.; Malorny, B. Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011–2018) in Germany. Front. Microbiol. 2020, 11, 80.
- Li, J.; Shi, X.; Yin, W.; Wang, Y.; Shen, Z.; Ding, S.; Wang, S. A multiplex SYBR green real-time PCR assay for the detection of three colistin resistance genes from cultured bacteria, feces, and environment samples. Front. Microbiol. 2017, 8, 2078.
- Neumann, B.; Rackwitz, W.; Hunfeld, K.P.; Fuchs, S.; Werner, G.; Pfeifer, Y. Genome sequences of two clinical Escherichia coli isolates harboring the novel colistin-resistance gene variants mcr-1.26 and mcr-1.27. Gut Pathog. 2020, 12, 40.
- Nicolas, I.; Bordeau, V.; Bondon, A.; Baudy-Floc’h, M.; Felden, B. Novel antibiotics effective against gram-positive and -negative multi-resistant bacteria with limited resistance. PLoS Biol. 2019, 17, e3000337.
- Flament-Simon, S.C.; de Toro, M.; Mora, A.; García, V.; García-Meniño, I.; Díaz-Jiménez, D.; Herrera, A.; Blanco, J. Whole genome sequencing and characteristics of mcr-1-harboring plasmids of porcine Escherichia coli isolates belonging to the high-risk clone O25b:H4-ST131 clade B. Front. Microbiol. 2020, 11, 387.




