1. Global Dissemination of mcr among Different Bacteria in Different Environments
It is believed that sporadic outbreaks of
mcr occurred in Chinese food-producing livestock in 1980
[1][17]. Since that time,
mcr-1-carrying bacterial strains have been reported in several countries among five of the seven continents across the globe
[1][2][3][4][5][6][17,25,43,104,105,106] including China
[2][25], India
[7][107], Pakistan
[8][108], Vietnam
[9][109], Laos
[10][110], USA
[11][111], Italy
[12][112], and Japan
[13][79].
The transmission of
mcr genes carrying pathogens could occur from animals to humans via direct contact with food animals and pets
[14][15][16][113,114,115]. Also, reservoirs for
mcr-1-carrying bacteria have been identified in public beaches
[17][116], hospital sewage, wastewater treatment plants
[18][19][117,118], rivers
[16][115], and water wells in rural areas
[20][119], as well as from houseflies and blowflies
[21][120]. Although data from some studies suggests that flies might be intermediate vectors for transmission of
mcr-1-containing bacteria between companion animals and humans
[22][121], the exact route for the spread of
mcr-1 and the bacteria carrying
mcr-1 needs more thorough investigation.
Several species of
Enterobacteriaceae possess
mcr-1, such as
E. coli where the gene is carried on IncI2 and IncX4 plasmids
[23][122],
Enterobacter aerogenes on an IncX4 plasmid
[24][123],
E. cloacae on an IncFI plasmid
[24][123],
Cronobacter sakazakii on an IncB/O plasmid
[25][124],
Citrobacter freundii on an IncHI2 plasmid
[26][125],
C. braakii on an IncI2-type plasmid,
K. pneumoniae on an IncX4 plasmid
[27][126],
Salmonella enterica on IncHI2-like plasmids
[28][127],
Shigella sonnei on IncHI2-like plasmids
[29][128], and
Raoultella ornithinolytican on an IncHI2 plasmid
[30][129]. Also,
mcr-1 variants have been identified in strains co-harboring
blaNDM-5 that confers carbapenem resistance to
E. coli [8][108]. The
mcr-1.1 gene has been found in the chromosome of
E. coli and plasmid p16BU137 of
K. pneumoniae from environmental isolates in China
[31][76]. Further details of recently discovered
mcr variants and their respective transposons and plasmids are given in
Table 1.
Table 1.
The evolutionary divergence among
mcr
variants (
mcr-1
to
mcr-10
) (a score of 1 indicates no divergence between variants; a score of 0 indicates complete divergence).
In Australia, colistin resistance was reported among poultry isolates of
Aeromonas hydrophila,
Alcaligenes faecalis,
Myroides odoratus,
Hafnia paralvei, and
Pseudochrobactrum spp. from a chicken processing unit in the state of Victoria
[32][130]. Furthermore,
mcr-1 was found in association with incompatibility group IncI2 plasmids from isolates in the state of New South Wales (NSW)
[33][131], and
mcr-1.1 has been detected in
E. coli [34][132]. Similarly,
mcr-
1.1 and
mcr-3 were found among MDR isolates of
Salmonella enterica 4 from human and animal sources in NSW
[34][35][132,133]. An evolutionary analysis of multiple drug-resistant
Salmonella enterica serovar 4 indicated that the spread of the
mcr-3 variant in lineages 1 and 3 was associated with overseas travel to Southeast Asia
[36][84]. Lineage 1 included
mcr-3.1- and
blaCTX-M-55-positive isolates of
Salmonella enterica sequence type 34 from Europe and Asia that were resistant to colistin and third-generation cephalosporins
[36][37][81,84]. Whilst
mcr-3.2 in lineage 3 was associated with IncHI2 pST3 and IncAC plasmids, wherein the colistin resistance genes were part of
dgkA (diacylglycerol kinase)
[36][38][84,134], which is a small transposable unit associated with IS elements circularized and integrated into
Enterobacterales genomes
[39][80].
2. Evolution of mcr Gene Variants from mcr-1 to mcr-10
In the current study, the phylogeny among mcr variants was determined using Molecular Evolutionary Genetics Analysis (MEGA 11) and is shown in Table 1. This shows the pair-end number of substitutions between mcr-1 and mcr-10, with the number of base differences per site indicated. An estimate of evolutionary divergence between the sequences of mcr-1 and mcr-10.1 was performed using MEGA 11. Overall, the average divergence among mcr ranged from 52 ± 20% for mcr-2 compared to all others to 69 ± 4% for mcr-8.
Moreover, phytogenic analysis of
mcr-3 also demonstrated that most occurred and evolved among
Aeromonas species. This suggested the origin of
mcr-
3 was
Aeromonas species with gradual evolution and transmission of
mcr-3 variants to
E. coli and
K. pneumoniae, while other
mcr gradually evolved among
E. coli and
K. pneumoniae. Interestingly, after the emergence of
mcr-
4, the identification of
mcr-4.3 in
A. baumannii represented a gradual evolution of
A. baumannii against colistin with a distinct type of
mcr gene in the form of a novel plasmid carrying
mcr-
4.3 [40][135].
The analysis of evolutionary probabilities in
mcr variants used a previously described method
[41][136] using modified evolutionary probabilities (EPs)
[42][137]. A user-specified tree topology was analyzed using the maximum likelihood method and the general time reversible model
[43][138]. The evolutionary time depths used in the EP calculation can be obtained using the real-time
[44][139] method. This analysis involved using the 10 nucleotide sequences of
mcr. Codon positions included the first + second + third plus the noncoding positions. All positions containing gaps and missing data were eliminated (complete deletion option). The results, which represent the number of base differences per site for each
mcr variant, are depicted in (
Figure 15).
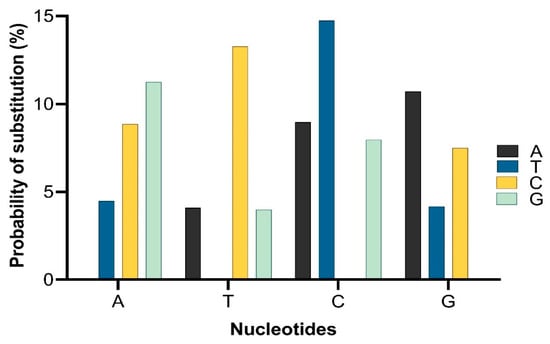
Figure 15. The probability of substitution of one base for another base. Substitution patterns and rates were estimated using the general time reversible model
[45][1]. The maximum log-likelihood for this computation was 2655.269. This analysis involved all 10 nucleotide sequences of
mcr. Codon positions included were 1st + 2nd + 3rd + noncoding. All positions containing gaps and missing data were eliminated (complete deletion option).
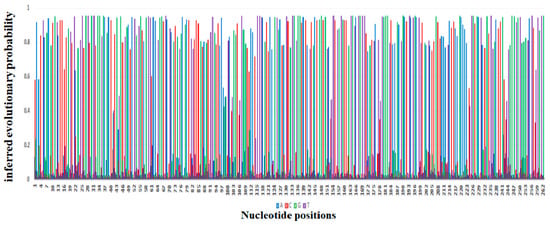
The probability of substitution of nucleotides to mcr-1 is demonstrated in Figure 15, which shows that the most likely substitution of adenine was with guanine (12%), of thymine was with cytosine (15%), of cytosine was with thymine (15%), and of guanine was with adenine (11%). The positions of substitution of nucleotides (A, T, G, and C from position 1 to 262 of different sites) for mcr-1 (E. coli strain ZZ1409 KU886144) are shown in Figure 26, respectively. In terms of positioning, cytosine (C) is predominately present at positions 1 to 257, followed by adenine (A) from positions 1 to 253, guanine (G) from positions 1 to 261, and thymine (T) from positions 5 to 261. In terms of probability and position of substitution, guanine was mostly likely to be present at position 27 with a probability of 0.95, and least likely to be present at position 28 with a probability of substitution of 0.007; thymine was most likely to be present at position 30 with a probability of 0.95 and least likely to be present at position 28 with a probability of 0.007; adenine was most likely to be present at position 220 with a probability of 0.94 and least likely to be present at position 27 with a probability of 0.007; cytosine was most likely to be present at position 160 with a probability of 0.93 and least likely to be present at position 262 with a probability of 0.014.
Figure 26.
Depiction of the evolutionary probabilities of nucleotide substitution with respect to positions 1 to 262 for
mcr-1
in
Escherichia coli
strain ZZ1409 KU886144.
The Processes and Molecular Vehicles Responsible for the Transmission of mcr Variants
Studies have comprehensively analyzed the genetic environments of
mcr-carrying genomes using bioinformatics tools such as Geneious R8
[46][140] and ISfinder software
[47][141] to demonstrate the insertion of
mcr variants. The structures of recently reported insertion sequences and the names of their associated transposons are given in
Table 2.
Full genome sequencing and analysis for identification of replication origin (
oriC) in
mcr-1-harboring plasmids from colistin-resistant isolates have identified a novel hybrid IncI2/IncFIB plasmid pGD17-2
[48][142]. Moreover, the co-occurrence of pGD17-2 with plasmids pGD65-3, IncI2, and pGD65-5, IncX4 has been reported in a single drug-resistant isolate (GD65), and this co-occurrence might promote the dissemination of
mcr-1 under environmental selection pressure
[48][142].
mcr-1 and other clinically significant resistant genes such as extended-spectrum β-lactamase (ESBL)
blaCTX-8 and
blaCTX-M-1 are related to globally identified sequence types ST10, ST46, and ST1638 in pathogenic strains of
E. coli responsible for infections in humans and animals
[49][50][51][143,144,145].
E. coli ST10 stains carrying
mcr-1 have been isolated from water at a public beach in the USA where the same ST10 strain had been isolated from an infected migratory Magellanic penguin with pododermatitis
[49][143], suggesting that the ST10 strains carrying
mcr-1 can disseminate in the marine environment.
E. coli mcr-1-positive environmental isolates have been isolated from German swine farms
[52][146] and in diseased food animals in China
[53][147], Italy, and France
[54][148]. A plastidome analysis of
mcr-
1 of
Enterobacterales human isolates suggested that the spread of
mcr-1 among commensals such as
K. pneumoniae,
E. coli, and other clinical isolates could be facilitated by various promiscuous diverse plasmids
[55][149].
Insertion sequences (ISs) or integrons can also facilitate the spread of
mcr. An analysis of
mcr-1 from various sources using whole genome sequencing supported a single
mcr-1 mobilization event in IS
Apl1-mcr-1-orf-IS
Apl1 transposon
[56][150]. This transposon has been immobilized on different plasmids such as IncI2, IncHI2, and IncX4
[57][151]. Plasmids pGD65-3, IncI2, and pGD65-5, IncX4 contain two insertion sequences, IS
Ecp1 and IS
Apl1, that facilitate the mobilization of
mcr-1 [48][142]. The insertion sequence IS
Apl1, which originated in
Actinobacillus pleuropneumoniae, is located upstream of
mcr-1 in the IncI2-type
mcr-1-harboring plasmid Phnshp45
[58][59][60][74,152,153]. However, the IS
Apl1 element is not always found associated with
mcr-1 on most IncX4 plasmids
[59][60][61][152,153,154]. A reason for this may be that the translocation of an
mcr-1-pap2 element by integration of an IS
Apl1 cassette (a member of the IS
30 family)
[38][59][134,152] into plasmids such as pMCR1-IncI2, and pMCR1-IncX4 may induce the formation of circular intermediates by recognizing inverted repeat sequences, which ultimately results in loss of IS
Apl1 after integration of
mcr-1 [38][62][63][134,155,156].
The
mcr-2 gene is not associated with IS
Apl1, but there are two IS
1595-like insertion sequences predicted to surround
mcr-2 in the IncX4 plasmid pKP37-BE
[64][157]. The short IS
1595-like element carries a transposase gene flanked by two inverted repeats surrounding
mcr-2. This transposase-encoding gene is similar (75% identity) to a fragment found in
Moraxella bovoculi strain 58069, which suggests the origin of
mcr-2 was from
M. bovoculi [62][155]. The occurrence of duplicate target sites adjacent to a spacer sequence suggests that the spacer sequence is the most probable hot site in IncX4 plasmids for integration and transposition of
mcr-2 variants
[65][158]. Transfer of
mcr-2 can occur through IS1595-containing transposons
[62][63][65][66][155,156,158,159].
Table 2.
Recently reported insertion sequences and transposon elements associated with
mcr
genes transmission.
3. Methods for Detecting Polymyxin Resistance
As resistance to polymyxins is being reported frequently among different bacterial isolates from humans, animals, and the environment, affordable, accessible, and efficient diagnostic approaches are needed. The phenotypic determination of colistin-resistant isolates can be made by growing on media such as CHROMagar COL- APSE
[79][171], SuperPolymyxin™
[80][172], and LBJMR
[81][173], as well as using commercial automated MIC-determining instruments such as BD Phoenix, MicroScan, Vitek 2
[82][174], MICRONAUT- S
[83][175], and Sensititre
[84][176]. The rapid polymyxin NP test and its modifications
[85][177], colispot
[86][178] colistin MAC test
[87][179], MIC Test Strip, MICRONAUT-MIC Strip
[88][180], the UMIC System
[89][181], and Sensitest Colistin
[84][176] can also be used
[82][174]. Eazyplex SuperBug kit
[90][182] and Taqman/SYBR Green real-time PCR assays have been used for molecular identification of
mcr genes that have yielded 100% specificity and sensitivity with a rapid turnaround time (<3 h)
[91][183]. More advanced molecular techniques such as multi-loop-mediated isothermal amplification (multi-LAMP) assays can also be used for rapid detection of
mcr genes
[92][184]. Based on cost, sensitivity and specificity, turnaround time, and the skills required to perform the test, the use of culture media or the Rapid Polymyxin Nordmann–Poirel (RPNP) test are recommended for low-resourced laboratories, while Multiplex PCR or Taqman/SYBR Green real-time PCR assays along with RPNP or novel culture media are applicable for well-resourced laboratories
[93][94][185,186].
To study the evolution in
mcr-positive bacterial strains, different sequencing techniques can be used including Sanger sequencing and the identification of single nucleotide polymorphisms
[95][187] for mutational analysis or identification of new
mcr- variant(s)
[96][188]. For detailed studies of intrinsic determinants of resistance, whole genome sequencing (WGS)
[97][189], nanopore sequencing, and transposon-directed insertion site sequencing
[72][165] can give insights into the interactions of genetic elements associated with polymyxins resistance. To study coevolution among pairs of
mcr or multiple
mcr elements within a single bacterial cell,
mcr-coevolution assays could be used
[72][165].