
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ki Hyun Nam | -- | 2261 | 2023-11-15 07:37:56 | | | |
| 2 | Peter Tang | + 2 word(s) | 2263 | 2023-11-15 09:46:32 | | |
Video Upload Options
Endo-1,4-β-xylanases belonging to the glycoside hydrolase (GH) 11 family hydrolyze the β-1,4-glycosidic linkages in the xylan backbone to convert polymeric xylan into xylooligosaccharides. GH11 xylanases play an essential role in sugar metabolism and are one of the most widely used enzymes in various industries, such as pulp and paper, food and feed, biorefinery, textile, and pharmaceutical industries. pH is a crucial factor influencing the biochemical properties of GH11 xylanase and its application in bioprocessing. For the optimal pH shifting of GH11 xylanase in industrial applications, various protein engineering studies using directed evolution, rational engineering, and in silico approaches have been adopted.
1. Introduction
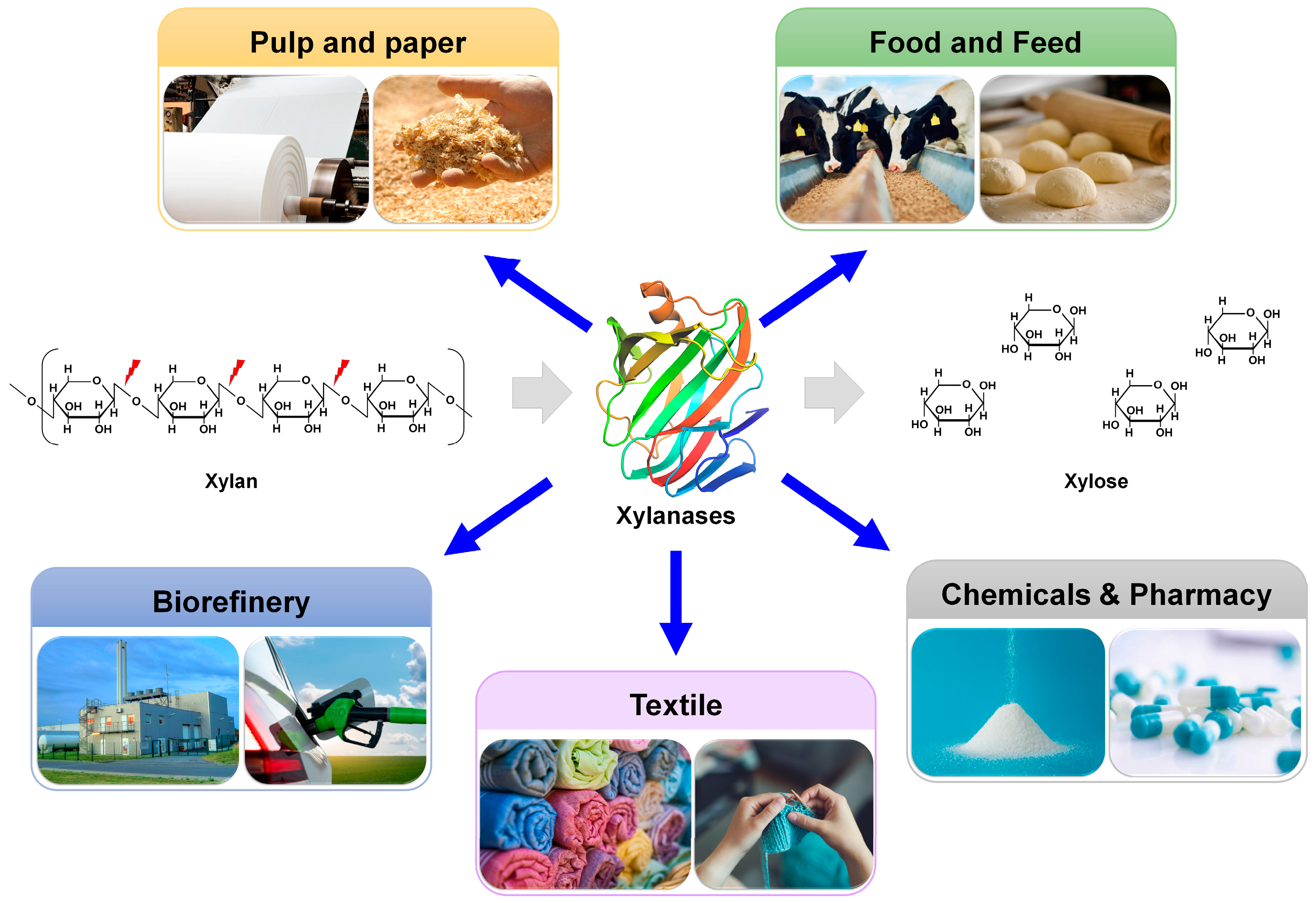
2. Functions and Structures of GH11 Xylanases
2.1. Catalytic Mechanisms
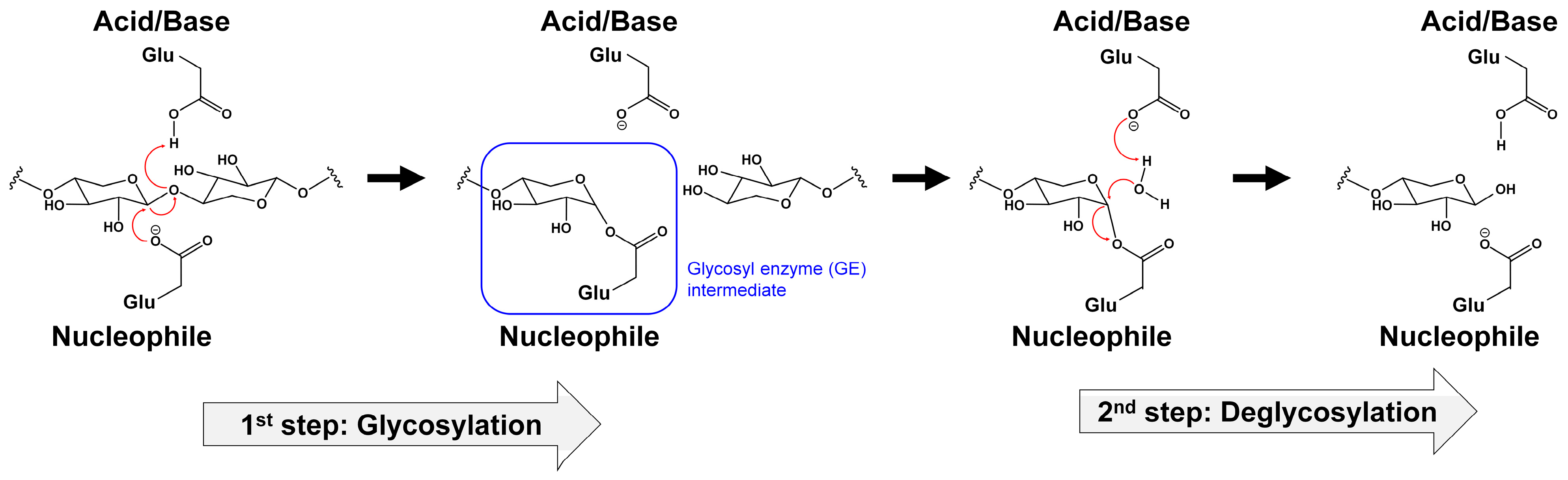
2.2. Structures
|
Accession No. |
Source Organism |
GENE NAME |
PDB Code (Complexed Ligand, Mutation) |
|---|---|---|---|
|
P36217 |
Trichoderma reesei (Hypocrea jecorina) or Trichoderma reesei RUT C-30 |
Endo-1,4-β-xylanase 2 (xyn2) |
1ENX, 1XYN, 1XYO, 1XYP, 1RED (4,5-epoxypentyl beta-D-xyloside), 1REE (3,4-epoxybutyl beta-D-xyloside), 1REF (2,3-epoxypropyl beta-D-xyloside), 2D97, 2D98, 2DFB, 2DFC, 3LGR (2,6-pyridinedicarboxylic acid), 4HK8 (xylohexaose), 4HK9 (xylotriose), 4HKL, 4HKO, 4HKW, 4S2D (MES), 4S2F, 4S2G, 4S2H, 4XQ4, 4XQD, 4XQW, 4XPV, 5K7P, 5ZF3 (xylotriose), 5ZH0, 5ZH9 (Y88F), 5ZII (xylotriose, Y88F), 5ZIW (Y77F), 5ZKZ (xylotriose, Y77F), 5ZO0, 6JUG (xylotriose), 6JWB (xylotriose), 6JXL (xylotriose), 6JZP (xylotriose), 6K9O, 6K9R (xylotriose), 6KW9 (xylotriose), 6KWD (xylotriose), 6KWF (xylotriose), 6KWG (xylotriose), 6K9W (xylotriose), 6KWC, 6K9X (xylotriose), 6KVV (xylotriose), 6KWE, 6KWH (xylotriose) |
|
P09850 |
Niallia circulans (Bacillus circulans) |
Endo-1,4-β-xylanase (xlnA) |
2BVV, 1BVV, 1C5H, 1C5I, 1HV0, 1HV1, 3VZJ, 3VZK, 3VZL, 3VZM (2-deoxy-2-fluoro-xylobiose), 3VZN (2-deoxy-2-fluoro-xylobiose), 3VZO (2-deoxy-2-fluoro-xylobiose) |
|
P18429 |
Bacillus subtilis (strain 168) |
Endo-1,4-β-xylanase A (xynA) |
1XXN, 2DCY, 2DCZ, 2B42 (Triticum xylanase inhibitor-IA), 2B45, 2B46, 2QZ3 (xylotetraose), 2Z79, 3HD8 (Triticum aestivum xylanase inhibitor-IIA), 3EXU, |
|
A0A7G1MBT0 |
Streptomyces olivaceoviridis (Streptomyces corchorusii) |
Endo-1,4-β-xylanase |
7DFM, 7DFN (α -L-arabinofuranosyl xylotetraose), 7DFO (4-O-methyl-α-D-glucuronopyranosyl xylotetraose) |
|
P55329 |
Aspergillus niger |
Endo-1,4-β-xylanase A (xynA) |
1T6G, 2QZ2 (xylopentaose), 6QE8 (xylobiose epoxide) |

3. Engineering of GH11 Xylanases for Optimal pH Shifting
|
Enzyme (Wild-Type) |
Source |
pH Optimum/Stability |
Condition |
Mutation Approach |
Reference |
|---|---|---|---|---|---|
|
XynA |
Bacillus subtilis |
pH 6 → 6.5 |
80 °C |
DE (epPCR and DNA shuffling) |
[44] |
|
CFXyl3 |
Cellulomonas flavigena |
pH 7 → 6 |
55 °C |
RD |
[45] |
|
Xyn5 |
Dictyoglomus thermophilum |
pH 7 → 6 |
65 °C, alkali-pretreated rice straw |
in silico analysis |
[10] |
|
TlXynA |
Thermomyces lanuginosus |
acid tolerance alkali tolerance |
65 °C, |
RD |
[46] |
|
BaxA |
Bacillus amyloliquefaciens |
pH 6 (broad pH spectrum) → 7 (narrow pH spectrum) |
50 °C |
DE (EPTD-PCR) |
[47] |
|
BCX |
Bacillus circulans |
pH 6.5 → 5–6 |
50 °C |
RD |
[18] |
|
XynB |
Thermotoga maritima |
pH 5.1 → 5.5, overall pH shift upwards by 0.5 |
90 °C |
RD |
[48] |
|
CbX-CD |
Caldicellulosiruptor bescii |
pH 6.5 → 5.0 |
70 °C |
RD Biomathematics and biostatistics |
[36] |
|
XynB |
Aspergillus niger |
pH 5.0 → 5.5 |
50 °C |
Biomathematics RD |
[31] |
References
- Bornscheuer, U.; Buchholz, K.; Seibel, J. Enzymatic Degradation of (Ligno)cellulose. Angew. Chem. Int. Ed. 2014, 53, 10876–10893.
- Peng, F.; Peng, P.; Xu, F.; Sun, R.-C. Fractional purification and bioconversion of hemicelluloses. Biotechnol. Adv. 2012, 30, 879–903.
- Ko, J.K.; Jung, M.W.; Kim, K.H.; Choi, I.-G. Optimal production of a novel endo-acting β-1,4-xylanase cloned from Saccharophagus degradans 2-40 into Escherichia coli BL21(DE3). New Biotechnol. 2009, 26, 157–164.
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238.
- Nguyen, S.T.C.; Freund, H.L.; Kasanjian, J.; Berlemont, R. Function, distribution, and annotation of characterized cellulases, xylanases, and chitinases from CAZy. Appl. Microbiol. Biotechnol. 2018, 102, 1629–1637.
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23.
- Paës, G.; Berrin, J.-G.; Beaugrand, J. GH11 xylanases: Structure/function/properties relationships and applications. Biotechnol. Adv. 2012, 30, 564–592.
- Nam, K.H.; Park, S.; Park, J. Preliminary XFEL data from spontaneously grown endo-1,4-β-xylanase crystals from Hypocrea virens. Acta Crystallogr. F Struct. Biol. Commun. 2022, 78, 226–231.
- Kim, I.J.; Kim, S.R.; Kim, K.H.; Bornscheuer, U.T.; Nam, K.H. Characterization and structural analysis of the endo-1,4-β-xylanase GH11 from the hemicellulose-degrading Thermoanaerobacterium saccharolyticum useful for lignocellulose saccharification. Sci. Rep. 2023, 13, 17332.
- Talens-Perales, D.; Sánchez-Torres, P.; Marín-Navarro, J.; Polaina, J. In silico screening and experimental analysis of family GH11 xylanases for applications under conditions of alkaline pH and high temperature. Biotechnol. Biofuels 2020, 13, 198.
- Walia, A.; Guleria, S.; Mehta, P.; Chauhan, A.; Parkash, J. Microbial xylanases and their industrial application in pulp and paper biobleaching: A review. 3 Biotech 2017, 7, 11.
- Sharma, N. Microbial xylanases and their industrial applications as well as future perspectives: A review. Glob. J. Biol. Agric. Health Sci. 2017, 6, 5–12.
- Viikari, L.; Kantelinen, A.; Sundquist, J.; Linko, M. Xylanases in bleaching: From an idea to the industry. FEMS Microbiol. Rev. 1994, 13, 335–350.
- Chen, C.-C.; Ko, T.-P.; Huang, J.-W.; Guo, R.-T. Heat- and Alkaline-Stable Xylanases: Application, Protein Structure and Engineering. ChemBioEng Rev. 2015, 2, 95–106.
- Vahjen, W.; Osswald, T.; Schäfer, K.; Simon, O. Comparison of a xylanase and a complex of non starch polysaccharide-degrading enzymes with regard to performance and bacterial metabolism in weaned piglets. Arch. Anim. Nutr. 2007, 61, 90–102.
- Lynd, L.R.; Laser, M.S.; Bransby, D.; Dale, B.E.; Davison, B.; Hamilton, R.; Himmel, M.; Keller, M.; McMillan, J.D.; Sheehan, J.; et al. How biotech can transform biofuels. Nat. Biotechnol. 2008, 26, 169–172.
- Sun, L.; Lee, J.W.; Yook, S.; Lane, S.; Sun, Z.; Kim, S.R.; Jin, Y.-S. Complete and efficient conversion of plant cell wall hemicellulose into high-value bioproducts by engineered yeast. Nat. Commun. 2021, 12, 4975.
- Pokhrel, S.; Joo, J.C.; Yoo, Y.J. Shifting the optimum pH of Bacillus circulans xylanase towards acidic side by introducing arginine. Biotechnol. Bioprocess Eng. 2013, 18, 35–42.
- Bhardwaj, N.; Kumar, B.; Verma, P. A detailed overview of xylanases: An emerging biomolecule for current and future prospective. Bioresour. Bioprocess. 2019, 6, 40.
- Hasunuma, T.; Kondo, A. Development of yeast cell factories for consolidated bioprocessing of lignocellulose to bioethanol through cell surface engineering. Biotechnol. Adv. 2012, 30, 1207–1218.
- Bornscheuer, U.T.; Huisman, G.W.; Kazlauskas, R.J.; Lutz, S.; Moore, J.C.; Robins, K. Engineering the third wave of biocatalysis. Nature 2012, 485, 185–194.
- Kan, S.B.J.; Lewis, R.D.; Chen, K.; Arnold, F.H. Directed evolution of cytochrome c for carbon–silicon bond formation: Bringing silicon to life. Science 2016, 354, 1048–1051.
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589.
- Dauparas, J.; Anishchenko, I.; Bennett, N.; Bai, H.; Ragotte, R.J.; Milles, L.F.; Wicky, B.I.M.; Courbet, A.; de Haas, R.J.; Bethel, N.; et al. Robust deep learning–based protein sequence design using ProteinMPNN. Science 2022, 378, 49–56.
- You, C.; Huang, Q.; Xue, H.; Xu, Y.; Lu, H. Potential hydrophobic interaction between two cysteines in interior hydrophobic region improves thermostability of a family 11 xylanase from Neocallimastix Patriciarum. Biotechnol. Bioeng. 2010, 105, 861–870.
- Zhang, Z.-G.; Yi, Z.-L.; Pei, X.-Q.; Wu, Z.-L. Improving the thermostability of Geobacillus stearothermophilus xylanase XT6 by directed evolution and site-directed mutagenesis. Bioresour. Technol. 2010, 101, 9272–9278.
- Song, L.; Siguier, B.; Dumon, C.; Bozonnet, S.; O’Donohue, M.J. Engineering better biomass-degrading ability into a GH11 xylanase using a directed evolution strategy. Biotechnol. Biofuels 2012, 5, 3.
- Xiang, L.; Wang, M.; Wu, L.; Lu, Z.; Tang, J.; Zhou, J.; Huang, W.; Zhang, G. Structural insights into xylanase mutant 254RL1 for improved activity and lower pH optimum. Enzyme Microb. Technol. 2021, 147, 109786.
- Cheng, Y.-S.; Chen, C.-C.; Huang, J.-W.; Ko, T.-P.; Huang, Z.; Guo, R.-T. Improving the catalytic performance of a GH11 xylanase by rational protein engineering. Appl. Microbiol. Biotechnol. 2015, 99, 9503–9510.
- Kumar, V.; Dangi, A.K.; Shukla, P. Engineering thermostable microbial xylanases toward its industrial applications. Mol. Biotechnol. 2018, 60, 226–235.
- Zhao, L.; Zhang, X.; Xie, J.; Li, F. Improvement of the Optimum pH of Aspergillus niger Xylanase towards an Alkaline pH by Site-Directed Mutagenesis. J. Microbiol. Biotechnol. 2015, 25, 11–17.
- Qiu, J.; Han, H.; Sun, B.; Chen, L.; Yu, C.; Peng, R.; Yao, Q. Residue mutations of xylanase in Aspergillus kawachii alter its optimum pH. Microbiol. Res. 2016, 182, 1–7.
- Yang, J.H.; Park, J.Y.; Kim, S.H.; Yoo, Y.J. Shifting pH optimum of Bacillus circulans xylanase based on molecular modeling. J. Biotechnol. 2008, 133, 294–300.
- Joshi, M.D.; Sidhu, G.; Pot, I.; Brayer, G.D.; Withers, S.G.; McIntosh, L.P. Hydrogen bonding and catalysis: A novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. J. Mol. Biol. 2000, 299, 255–279.
- Permyakov, E.A.; Bai, W.; Zhou, C.; Zhao, Y.; Wang, Q.; Ma, Y. Structural insight into and mutational analysis of family 11 xylanases: Implications for mechanisms of higher pH catalytic adaptation. PLoS ONE 2015, 10, e0132834.
- Ma, F.; Xie, Y.; Luo, M.; Wang, S.; Hu, Y.; Liu, Y.; Feng, Y.; Yang, G.-Y. Sequence homolog-based molecular engineering for shifting the enzymatic pH optimum. Synth. Syst. Biotechnol. 2016, 1, 195–206.
- Sürmeli, Y.; Şanlı-Mohamed, G. Engineering of xylanases for the development of biotechnologically important characteristics. Biotechnol. Bioeng. 2023, 120, 1171–1188.
- Ardèvol, A.; Rovira, C. Reaction mechanisms in carbohydrate-active enzymes: Glycoside hydrolases and glycosyltransferases. Insights from ab Initio quantum mechanics/molecular mechanics dynamic simulations. J. Am. Chem. Soc. 2015, 137, 7528–7547.
- McCarter, J.D.; Stephen Withers, G. Mechanisms of enzymatic glycoside hydrolysis. Curr. Opin. Struc. Biol. 1994, 4, 885–892.
- Sinnott, M.L. Catalytic mechanism of enzymic glycosyl transfer. Chem. Rev. 2002, 90, 1171–1202.
- Nam, K.H.; Lee, W.H.; Rhee, K.H.; Hwang, K.Y. Structural characterization of the bifunctional glucanase-xylanase CelM2 reveals the metal effect and substrate-binding moiety. Biochem. Biophys. Res. Commun. 2010, 391, 1726–1730.
- Ishida, T.; Parks, J.M.; Smith, J.C. Insight into the catalytic mechanism of GH11 xylanase: Computational analysis of substrate distortion based on a neutron structure. J. Am. Chem. Soc. 2020, 142, 17966–17980.
- Hutchison, C.A.; Phillips, S.; Edgell, M.H.; Gillam, S.; Jahnke, P.; Smith, M. Mutagenesis at a specific position in a DNA sequence. J. Biol. Chem. 1978, 253, 6551–6560.
- Ruller, R.; Deliberto, L.; Ferreira, T.L.; Ward, R.J. Thermostable variants of the recombinant xylanase a from Bacillus subtilis produced by directed evolution show reduced heat capacity changes. Proteins 2007, 70, 1280–1293.
- Tian, W.; Zhang, Z.; Yang, C.; Li, P.; Xiao, J.; Wang, R.; Du, P.; Li, N.; Wang, J. Engineering mesophilic GH11 xylanase from Cellulomonas flavigena by rational design of N-terminus substitution. Front. Bioeng. Biotechnol. 2022, 10, 1044291.
- Wu, X.; Zhang, Q.; Zhang, L.; Liu, S.; Chen, G.; Zhang, H.; Wang, L. Insights into the role of exposed surface charged residues in the alkali-tolerance of GH11 xylanase. Front. Microbiol. 2020, 11, 872.
- Xu, X.; Liu, M.-Q.; Huo, W.-K.; Dai, X.-J. Obtaining a mutant of Bacillus amyloliquefaciens xylanase A with improved catalytic activity by directed evolution. Enzyme Microb. Technol. 2016, 86, 59–66.
- Liu, L.; Wang, B.; Chen, H.; Wang, S.; Wang, M.; Zhang, S.; Song, A.; Shen, J.; Wu, K.; Jia, X. Rational pH-engineering of the thermostable xylanase based on computational model. Process Biochem. 2009, 44, 912–915.




