Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Frederick Kibenge | -- | 1558 | 2023-11-05 15:02:17 | | | |
| 2 | Peter Tang | Meta information modification | 1558 | 2023-11-06 02:50:05 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Godoy, M.; Oca, M.M.D.; Caro, D.; Pontigo, J.P.; Kibenge, M.; Kibenge, F. Influenza A Virus Subtypes in Chile. Encyclopedia. Available online: https://encyclopedia.pub/entry/51158 (accessed on 07 February 2026).
Godoy M, Oca MMD, Caro D, Pontigo JP, Kibenge M, Kibenge F. Influenza A Virus Subtypes in Chile. Encyclopedia. Available at: https://encyclopedia.pub/entry/51158. Accessed February 07, 2026.
Godoy, Marcos, Marco Montes De Oca, Diego Caro, Juan Pablo Pontigo, Molly Kibenge, Frederick Kibenge. "Influenza A Virus Subtypes in Chile" Encyclopedia, https://encyclopedia.pub/entry/51158 (accessed February 07, 2026).
Godoy, M., Oca, M.M.D., Caro, D., Pontigo, J.P., Kibenge, M., & Kibenge, F. (2023, November 05). Influenza A Virus Subtypes in Chile. In Encyclopedia. https://encyclopedia.pub/entry/51158
Godoy, Marcos, et al. "Influenza A Virus Subtypes in Chile." Encyclopedia. Web. 05 November, 2023.
Copy Citation
The influenza A virus (IAV) poses a significant global threat to public health and food security. Chile’s encounter with IAV began in 2002, with the highly pathogenic avian influenza (HPAI) H7N3 virus, derived from a unique South American low pathogenic avian influenza (LPAI) virus.
influenza A virus
avian influenza virus
poultry farms
1. Introduction
Influenza viruses belong to the Orthomyxoviridae family [1], and, along with the Amnoonviridae family [2], they constitute the order Articulavirales [3]. Members of this family possess a segmented negative-sense single-stranded RNA genome. The number of genome segments varies, depending on the genus: Orthomyxoviridae has 6–8 segments, while Amnoonviridae has 10. Influenza viruses are classified into four genera: Alphainfluenzavirus, Betainfluenzavirus, Gammainfluenzavirus, and Deltainfluenzavirus. Each genus has a single ratified species: influenza A virus (IAV), influenza B virus (IBV), influenza C virus (ICV), and influenza D virus (IDV), respectively [4]. The Orthomyxoviridae family includes not only influenza viruses but also other orthomyxoviruses categorized into different genera, such as Thogotovirus, Isavirus, Mykissvirus, Sardinovirus, and Quarajavirus [1][4][5][6]. Most recently, the reclassification of the genus Quarajavirus as the family Quaranjaviridae has been proposed. Additionally, a novel and divergent family, tentatively named Cnidenomoviridae, has been discovered in Cnidaria (including corals) and is now included in the order Articulavirales, bringing the total number of families in this order to four [7].
2. AIV H7N3
The first IAV disease occurrence in commercial poultry in Chile was in 2002 and was caused by HPAI H7N3 (Figure 1), characterized as lineage A/chicken/Chile/184240-4322/2002(H7N3), which had mutated from an LPAI virus [8]. The epicenter of the outbreak was a broiler breeder farm in the densely populated poultry region of San Antonio, V Region, Chile. The farm consisted of 27 poultry sheds housing birds ranging from 1 to 79 weeks in age, besides a hatchery [9]. Between April and May 2002, the farm experienced a clinical disease characterized by a relatively low mortality rate, a slight decline in egg production, and cases of salpingo-peritonitis. In certain cases, mortality was so sudden that no clinical signs were observable. The necropsy revealed cyanotic combs and wattles, as well as petechial hemorrhages in various organs, such as the muscles, heart, pancreas, and legs. Subcutaneous edema was also present. Subsequent surveillance activities identified a second outbreak occurring one week later at a turkey breeding farm owned by the same company. This farm, situated 4 km from the initial outbreak, consisted of eight pens that housed turkeys ranging in age from 6 to 59 weeks, besides a hatchery. The infection was confined to 25% of the sheds. Clinical signs predominantly affected the upper respiratory tract and were followed by a sudden increase in mortality. Cloacal swabs were collected, and an HPAI virus was isolated from two samples from this farm, which were identical to those obtained from the index case [9].
The initial identification of LPAI H7N3 was followed by the detection of HPAI H7N3, which was accompanied by increased mortality rates. Detailed analysis of the viral genomes has yielded valuable insights, revealing minimal genetic differences between the low and highly pathogenic strains, except for a significant alteration in the cleavage site of the HA protein. The LPAI H7N3 virus exhibited a cleavage site similar to that of other LPAI H7 viruses. In contrast, the HPAI H7N3 isolates displayed a 30-nucleotide insertion at this specific site. This insertion possibly resulted from recombination events between the HA and nucleoprotein genes of the LPAI H7N3, leading to an increase in pathogenicity. A comparative analysis of the full sequences of the eight gene segments confirmed that the Chilean viruses had a distinct nature vis-a-vis other AIVs, forming a unique clade exclusive to South America. These findings highlight the critical importance of the continuous monitoring and surveillance of AIVs. These viruses can undergo mutations that significantly increase their pathogenicity [10]. In addition, identifying a distinct viral clade specific to South America underscores the necessity for an understanding of and control strategies tailored to effectively managing AIV outbreaks in the region [11].
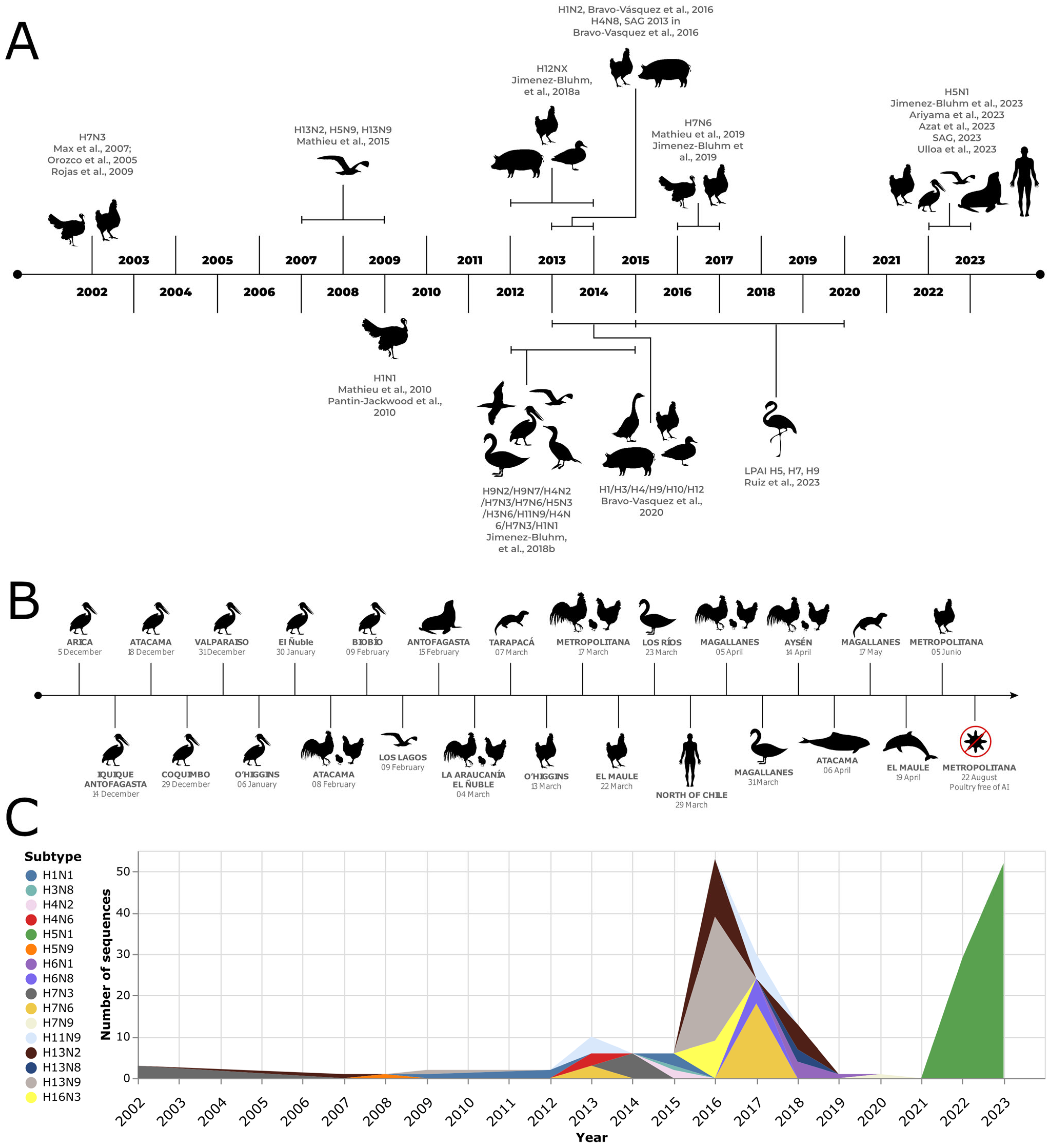
Figure 1. Influenza A virus outbreak in birds and non-human mammals in Chile. (A) Literature review derives a timeline of outbreaks [8][9][12][13][14][15][16][17][18][19][20][21][22][23][24][25][26][27]. (B) H5N1 cases were registered during late 2022 and 2023, based on data from the Chilean Agricultural and Livestock Service [24] and the National Fisheries and Aquaculture Service [28]. (C) Number of hemagglutinin sequences available from avian samples and classified by subtype per year in Chile. Only the three most abundant subtypes per year are shown. Sequences retrieved from the NCBI nucleotide database [29].
Interestingly, isolates obtained from the HPAI H7N3 outbreak in commercial poultry in Chile in May 2002 [9] shared similarity in some genes with an LPAI H7N3 virus isolated from a Cinnamon Teal (Anas cyanoptera) specimen in Bolivia in 2001, whose NA and matrix (M) genes shared the highest sequence similarity with North American AIV isolates. The identification of genetic similarities between AIV isolates from wild birds in North America and outbreaks in commercial poultry in Chile further emphasize the importance of the ongoing surveillance and understanding of AIVs in both wild and domestic bird populations [30].
3. Pandemic Influenza A (H1N1) 2009 (H1N1pdm09)
The pandemic influenza A (H1N1) 2009 (H1N1pdm09) virus, which caused the fourth influenza pandemic [31], emerged as a triple reassortant virus in pigs [32]. In animals, it was first recognized in Canada in May 2009, where it caused respiratory disease in pigs, and in Chile in June 2009, where it led to a significant decrease in egg production in turkeys [13]. The reverse zoonotic virus transmission to turkeys in 2009 remained somewhat a mystery. The artificial insemination of turkeys by infected staff has been proposed as an unusual but possible pathway, considering that, in modern turkey production, turkey hens are handled once a week for intrauterine insemination in order to produce fertile eggs [13]. During the outbreak in turkeys in Chile, there was a decrease in egg production and shell quality among turkey flocks on two farms (A and B) located in the Valparaiso region of Chile. The suspicion of AIV led to the collection of blood samples from the affected turkeys on 14 August 2009, for serological testing [14]. The agar gel immunodiffusion (AGID) assay detected IAV antibodies in 140 out of 227 turkeys sampled, with a higher proportion of positive cases in Farm A (80%) than in Farm B (32%) [14]. Consequently, the Chilean Agricultural and Livestock Service (SAG) implemented control measures, including quarantine, intensified biosecurity measures, epidemiological investigations, and postmortem examinations [14]. Sampling for the detection of viral RNA was conducted on 16 August 2009, on affected flocks, surrounding premises, and neighboring turkey farms. Two days later, the results showed that the infection was present only in the breeder turkeys from the initially affected flocks of Farms A and B. The real-time reverse transcription–quantitative polymerase chain reaction (RT-qPCR) identified RNA corresponding to the IAV M gene but not the H5 or H7 genes. Viral RNA was detected in some cloacal and tracheal swabs but not in the homogenates of turkey embryo lungs and tracheas. This ruled out vertical transmission. Necropsies revealed specific lesions in some birds, while others were recovering. Fecal samples from wild birds near Farm A tested negative for the influenza virus. Subtyping tests identified the virus subtype as H1N1. On 19 August 2009, SAG authorities coordinated with the Chilean Public Health Institute (ISP) to isolate the virus and sequence its genome. The viral sequences exhibited characteristics almost identical to the novel influenza A (H1N1) pdm09 virus [14], suggesting reverse zoonosis, namely the transmission of the influenza A (H1N1) pdm09 virus from humans to turkeys. The virus was classified as belonging to the A/turkey/Chile/28317-6504-3/2009(H1N1) lineage. Follow-up testing indicated that the virus was eliminated from turkeys within 2–4 weeks. During September and October, SAG implemented an RT-qPCR assay to detect the N1 gene of the influenza A (H1N1) pdm09 virus as the flock’s egg production gradually recovered. The most recent evidence of infection was obtained on 31 August 2009, indicating the successful eradication of the virus from the turkeys [14].
4. AIV H7N6
In December 2016, a turkey farm in the Valparaiso region of Chile was infected with LPAI H7N6. Two weeks later, another turkey farm 70 km north of the first site was also infected. The animals presented a variety of gross lesions, including bursitis, catarrhal to mucopurulent caseous sinusitis, tracheitis, caseous/purulent airsacculitis, polyserositis, pericarditis/hydropericardium, congestion, pulmonary edema, mucopurulent to caseous pneumonia, localized subcutaneous emphysema, mild splenomegaly, pancreatitis, petechiae, ecchymosis focused on the epicardium and coronary fat, fibrin-purulent pericarditis, pulmonary congestion/mucopurulent to caseous pneumonia, and pleuritis. On 28 January 2017, a backyard poultry farm also tested positive for AIV through the AGID test. However, the PCR test result was negative. Various measures, such as depopulation, zoning, animal movement control, and active surveillance, were implemented to contain the outbreak. The 2016–2017 Chilean LPAI outbreak in turkeys involved a group of viruses belonging to a monophyletic clade. These viruses share the closest common ancestor with two other viruses, A/yellow_billed_pintail/Chile/10/2014(H7N3) and A/yellow_billed_teal/Chile/9/2013(H7N6), collected from wild water birds in the same area. The A/turkey/Chile/2017(H7N6) LPAI virus is part of a native South American lineage. Phylogenetic analysis revealed a close relationship between the A/turkey/Chile/2017(H7N6) LPAI viruses and AIV found in wild aquatic birds in Chile and Bolivia, as well as the A/chicken/Chile 2002 virus, which caused the HPAI H7N3 outbreak in Chile. Based on the HA phylogeny, these South American AIVs formed a distinct cluster [15][16].
References
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H. Create a Megataxonomic Framework, Filling All Principal Taxonomic Ranks, for Realm Riboviria; ICTV: Montolivet, France, 2019; Unpublished.
- Turnbull, O.M.H.; Ortiz-Baez, A.S.; Eden, J.-S.; Shi, M.; Williamson, J.E.; Gaston, T.F.; Zhang, Y.-Z.; Holmes, E.C.; Geoghegan, J.L. Meta-Transcriptomic Identification of Divergent Amnoonviridae in Fish. Viruses 2020, 12, 1254.
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062.
- Parry, R.; Wille, M.; Turnbull, O.M.H.; Geoghegan, J.L.; Holmes, E.C. Divergent Influenza-Like Viruses of Amphibians and Fish Support an Ancient Evolutionary Association. Viruses 2020, 12, 1042.
- García-Sastre, A. Create One New Genus (Mykissvirus) Including One New Species (Mykissvirus tructae) (Articulavirales: Orthomyxoviridae). 2021. Available online: https://ictv.global/ictv/proposals/2021.022M.R.Orthomyxoviridae_1ngen_1nsp_Mykiss.zip (accessed on 5 June 2023).
- Mohr, P.G.; Godwin, S.E.; Morrison, R.N.; Carson, J.; Crane, M.S.t.J.; Moody, N.J.G. Create One New Genus (Sardinovirus) Including One New Species (Sardinovirus pilchardi) (Articulavirales: Orthomyxoviridae). 2021. Available online: https://ictv.global/ictv/proposals/2021.023M.R.Orthomyxoviridae_1ngen_1nsp_Sardino.zip (accessed on 2 June 2023).
- Petrone, M.E.; Parry, R.; Mifsud, J.C.O.; Van Brussel, K.; Vorhees, I.; Richards, Z.T.; Holmes, E.C. Evidence for an Aquatic Origin of Influenza Virus and the order Articulavirales. bioRxiv 2023. preprint.
- Orozco, C. The SPS Agreement and Crisis Management: The Chile–EU Avian Influenza Experience. Managing the Challenges of WTO Participation; Cambridge University Press: Cambridge, UK, 2005; pp. 150–166.
- Max, V.; Herrera, J.; Moreira, R.; Rojas, H. Avian Influenza in Chile: A Successful Experience. Avian Dis. 2007, 51, 363–365.
- Olsen, B.; Munster, V.J.; Wallensten, A.; Waldenström, J.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Global Patterns of Influenza A Virus in Wild Birds. Science 2006, 312, 384–388.
- Suarez, D.L.; Senne, D.A.; Banks, J.; Brown, I.H.; Essen, S.C.; Lee, C.-W.; Manvell, R.J.; Mathieu-Benson, C.; Moreno, V.; Pedersen, J.C.; et al. Recombination Resulting in Virulence Shift in Avian Influenza Outbreak, Chile. Emerg. Infect. Dis. 2004, 10, 693–699.
- Rojas Olavarría, H.; Moreira Zúñiga, R.; Mundial, B. Influenza Aviar en Chile 2002: Una Sinopsis; Instituto Interamericano de Cooperación para la Agricultura (IICA)—Banco Mundial (BM): Santiago, Chile, 2009; 32p.
- Pantin-Jackwood, M.; Wasilenko, J.L.; Spackman, E.; Suarez, D.L.; Swayne, D.E. Susceptibility of turkeys to pandemic-H1N1 virus by reproductive tract insemination. Virol. J. 2010, 7, 27.
- Mathieu, C.; Moreno, V.; Retamal, P.; Gonzalez, A.; Rivera, A.; Fuller, J.; Jara, C.; Lecocq, C.; Rojas, M.; García, A.; et al. Pandemic (H1N1) 2009 in Breeding Turkeys, Valparaiso, Chile. Emerg. Infect. Dis. 2010, 16, 709–711.
- Mathieu, C.; Gonzalez, A.; Garcia, A.; Johow, M.; Badia, C.; Jara, C.; Nuñez, P.; Neira, V.; Montiel, N.A.; Killian, M.L.; et al. H7N6 low pathogenic avian influenza outbreak in commercial turkey farms in Chile caused by a native South American Lineage. Transbound. Emerg. Dis. 2019, 68, 2–12.
- Jimenez-Bluhm, P.; Bravo-Vasquez, N.; Torchetti, M.K.; Killian, M.L.; Livingston, B.; Herrera, J.; Fuentes, M.; Schultz-Cherry, S.; Hamilton-West, C. Low pathogenic avian influenza (H7N6) virus causing an outbreak in commercial Turkey farms in Chile. Emerg. Microbes Infect. 2019, 8, 479–485.
- Jimenez-Bluhm, P.; Di Pillo, F.; Bahl, J.; Osorio, J.; Schultz-Cherry, S.; Hamilton-West, C. Circulation of influenza in backyard productive systems in central Chile and evidence of spillover from wild birds. Prev. Vet. Med. 2018, 153, 1–6.
- Bravo-Vasquez, N.; Di Pillo, F.; Lazo, A.; Jiménez-Bluhm, P.; Schultz-Cherry, S.; Hamilton-West, C. Presence of influenza viruses in backyard poultry and swine in El Yali wetland, Chile. Prev. Vet. Med. 2016, 134, 211–215.
- Bravo-Vasquez, N.; Baumberger, C.; Jimenez-Bluhm, P.; Di Pillo, F.; Lazo, A.; Sanhueza, J.; Schultz-Cherry, S.; Hamilton-West, C. Risk factors and spatial relative risk assessment for influenza A virus in poultry and swine in backyard production systems of central Chile. Vet. Med. Sci. 2020, 6, 518–526.
- Mathieu, C.; Moreno, V.; Pedersen, J.; Jeria, J.; Agredo, M.; Gutiérrez, C.; García, A.; Vásquez, M.; Avalos, P.; Retamal, P. Avian Influenza in wild birds from Chile, 2007–2009. Virus Res. 2015, 199, 42–45.
- Jiménez-Bluhm, P.; Karlsson, E.A.; Freiden, P.; Sharp, B.; Di Pillo, F.; Osorio, J.E.; Hamilton-West, C.; Schultz-Cherry, S. Wild birds in Chile Harbor diverse avian influenza A viruses. Emerg. Microbes Infect. 2018, 7, 1–4.
- Ruiz, S.; Galdames, P.; Baumberger, C.; Gonzalez, M.A.; Rojas, C.; Oyarzun, C.; Orozco, K.; Mattar, C.; Freiden, P.; Sharp, B.; et al. Remote Sensing and Ecological Variables Related to Influenza A Prevalence and Subtype Diversity in Wild Birds in the Lluta Wetland of Northern Chile. Viruses 2023, 15, 1241.
- Ariyama, N.; Pardo Roa, C.; Munoz, G.; Aguayo, C.; Avila, C.; Mathieu, C.; Brito, B.; Medina, R.; Johow, M.; Neira-Ramirez, V. Emergence and rapid dissemination of highly pathogenic avian influenza virus H5N1 clade 2.3.4.4b in wild birds, Chile. bioRxiv 2023. preprint.
- SAG (Agricultural and Livestock Service). Servicio Agrícola y Ganadero, Ministerio de Agricultura de Chile. Plataforma de Influenza Aviar. 2023. Available online: https://www.sag.gob.cl/ia (accessed on 25 May 2023).
- Jimenez-Bluhm, P.; Siegers, J.Y.; Tan, S.; Sharp, B.; Freiden, P.; Johow, M.; Orozco, K.; Ruiz, S.; Baumberger, C.; Galdames, P.; et al. Detection and phylogenetic analysis of highly pathogenic A/H5N1 avian influenza clade 2.3.4.4b virus in Chile, 2022. Emerg. Microbes Infect. 2023, 12, 2220569.
- Azat, C.; Alvarado-Rybak, M.; Aguilera, J.F.; Benavides, J.A. Spatio-Temporal Dynamics and Drivers of Highly Pathogenic Avian Influenza H5N1 in Chile. bioRxiv 2023. preprint.
- Ulloa, M.; Fernández, A.; Ariyama, N.; Colom-Rivero, A.; Rivera, C.; Nuñez, P.; Sanhueza, P.; Johow, M.; Araya, H.; Torres, J.C.; et al. Mass Mortality Event in South American Sea Lions (Otaria flavescens) Correlated to Highly Pathogenic Avian Influenza (HPAI) H5N1 Outbreak in Chile. In Veterinary Quarterly; Informa UK Limited: London, UK, 2023; pp. 1–13.
- SERNAPESCA. Servicio Nacional de Pesca. Plataforma de Influenza Aviar. 2023. Available online: http://www.sernapesca.cl/influenza-aviar (accessed on 7 September 2023).
- Nucleotide. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. 2004. Available online: https://www.ncbi.nlm.nih.gov/nuccore (accessed on 7 September 2023).
- Spackman, E.; McCracken, K.G.; Winker, K.; Swayne, D.E. H7N3 Avian Influenza Virus Found in a South American Wild Duck Is Related to the Chilean 2002 Poultry Outbreak, Contains Genes from Equine and North American Wild Bird Lineages, and Is Adapted to Domestic Turkeys. J. Virol. 2006, 80, 7760–7764.
- Kibenge, F.S.B. A One Health approach to mitigate the impact of influenza A virus (IAV) reverse zoonosis is by vaccinating humans and susceptible farmed and pet animals. Am. J. Vet. Res. 2023, 84, 1–9.
- Trifonov, V.; Khiabanian, H.; Rabadan, R. Geographic Dependence, Surveillance, and Origins of the 2009 Influenza A (H1N1) Virus. N. Engl. J. Med. 2009, 361, 115–119.
More
Information
Subjects:
Veterinary Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
550
Revisions:
2 times
(View History)
Update Date:
06 Nov 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




