Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Zahra Tabatabaei | -- | 1690 | 2023-10-16 11:43:32 | | | |
| 2 | Camila Xu | Meta information modification | 1690 | 2023-10-17 02:21:37 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tabatabaei, Z.; Wang, Y.; Colomer, A.; Oliver Moll, J.; Zhao, Z.; Naranjo, V. World-Wide Federated Content-Based Medical Image Retrieval. Encyclopedia. Available online: https://encyclopedia.pub/entry/50339 (accessed on 13 January 2026).
Tabatabaei Z, Wang Y, Colomer A, Oliver Moll J, Zhao Z, Naranjo V. World-Wide Federated Content-Based Medical Image Retrieval. Encyclopedia. Available at: https://encyclopedia.pub/entry/50339. Accessed January 13, 2026.
Tabatabaei, Zahra, Yuandou Wang, Adrián Colomer, Javier Oliver Moll, Zhiming Zhao, Valery Naranjo. "World-Wide Federated Content-Based Medical Image Retrieval" Encyclopedia, https://encyclopedia.pub/entry/50339 (accessed January 13, 2026).
Tabatabaei, Z., Wang, Y., Colomer, A., Oliver Moll, J., Zhao, Z., & Naranjo, V. (2023, October 16). World-Wide Federated Content-Based Medical Image Retrieval. In Encyclopedia. https://encyclopedia.pub/entry/50339
Tabatabaei, Zahra, et al. "World-Wide Federated Content-Based Medical Image Retrieval." Encyclopedia. Web. 16 October, 2023.
Copy Citation
Content-based medical image retrieval (CBMIR) is a recent DL-based methodology that allows pathologists a quick and precise search in previously diagnosed and treated cases. In CBMIR, image features, such as texture, shape, color, and intensity, are extracted from the query and data set; then, a similarity measure is applied to compare the query features with the features of the database.
breast cancer
content-based medical image retrieval (CBMIR)
convolutional auto-encoder (CAE)
1. Introduction
Breast cancer accounts for 25% of all cancers in women worldwide. According to the American Cancer Society, a woman is diagnosed with breast cancer in the world every 14 s. In the year 2020, approximately 2.3 million women were diagnosed with breast cancer globally, and 685,000 lost their lives due to it [1]. Histopathology is commonly used in the diagnosis and treatment of various diseases, including cancer. A biopsy, which is the removal of a small piece of tissue from the body, is usually required for histopathological examination [2]. Human error in histopathology refers to mistakes or inaccuracies made during the process of examining tissues or cells under a microscope [3]. Some examples of human errors in histopathology include sampling errors, processing errors, technical errors, interpretation errors, and reporting errors [4]. To minimize human errors in histopathology, it is essential to follow strict protocols and guidelines, perform regular quality control checks, and ensure that all personnel involved in the process are properly trained and competent [5]. The authors in [6] analyzed the accuracy of breast cancer diagnosis in 102 cases and found that there were diagnostic errors in 15.7% of cases. The most common types of errors were misclassification of tumor type and misinterpretation of pathology slides. Digital pathology could help pathologists to improve the accuracy and efficiency of cancer diagnosis, reduce the risk of errors, and enhance patient care.
Digital pathology is a technology that uses digital images of tissues and cells to aid in the diagnosis and management of diseases [7]. Deep learning (DL) has revolutionized computer-aided diagnosis (CAD) in digital pathology and has opened the door to improve cancer diagnosis while decreasing the pathologist’s workload [8].
Content-based medical image retrieval (CBMIR) is a recent DL-based methodology that allows pathologists a quick and precise search in previously diagnosed and treated cases [9]. In CBMIR, image features, such as texture, shape, color, and intensity, are extracted from the query and data set; then, a similarity measure is applied to compare the query features with the features of the database [10]. The retrieved images are ranked according to their similarity to the query image, and the most relevant images are displayed to the user.
To further illustrate the advantages and practicality of CBMIR in the field of histopathology and cancer diagnosis, consider a scenario where a patient is diagnosed with cancer, and grading it accurately poses a challenge for pathologists. In traditional cancer diagnosis methods, the pathologist would need to physically send the glass slide containing the tissue sample to another hospital, which could be located in a different city or even a different country. This process is not only expensive and time-consuming but also carries inherent risks, such as the loss or damage of the glass slide during transportation. Moreover, it adds additional stress to the patient’s already difficult situation.
By implementing world-wide content-based medical image retrieval (WWCBMIR), these challenges can be effectively addressed, and the process of a cancer diagnosis can be significantly expedited without compromising accuracy. Through the use of digital pathology, where whole-slide images (WSIs) are digitized and stored electronically, pathologists can access and analyze the images remotely [2]. The WWCBMIR enables pathologists to retrieve similar cases and relevant information from a vast database of histopathological images without the need for the physical transfer of slides. This approach not only reduces costs and saves time but also minimizes the potential risks associated with the transportation of delicate tissue samples. Figure 1 shows how a WWCBMIR can provide unprecedented access to K number of patches with the most similar patterns, allowing the pathologists to make a more confident diagnosis.
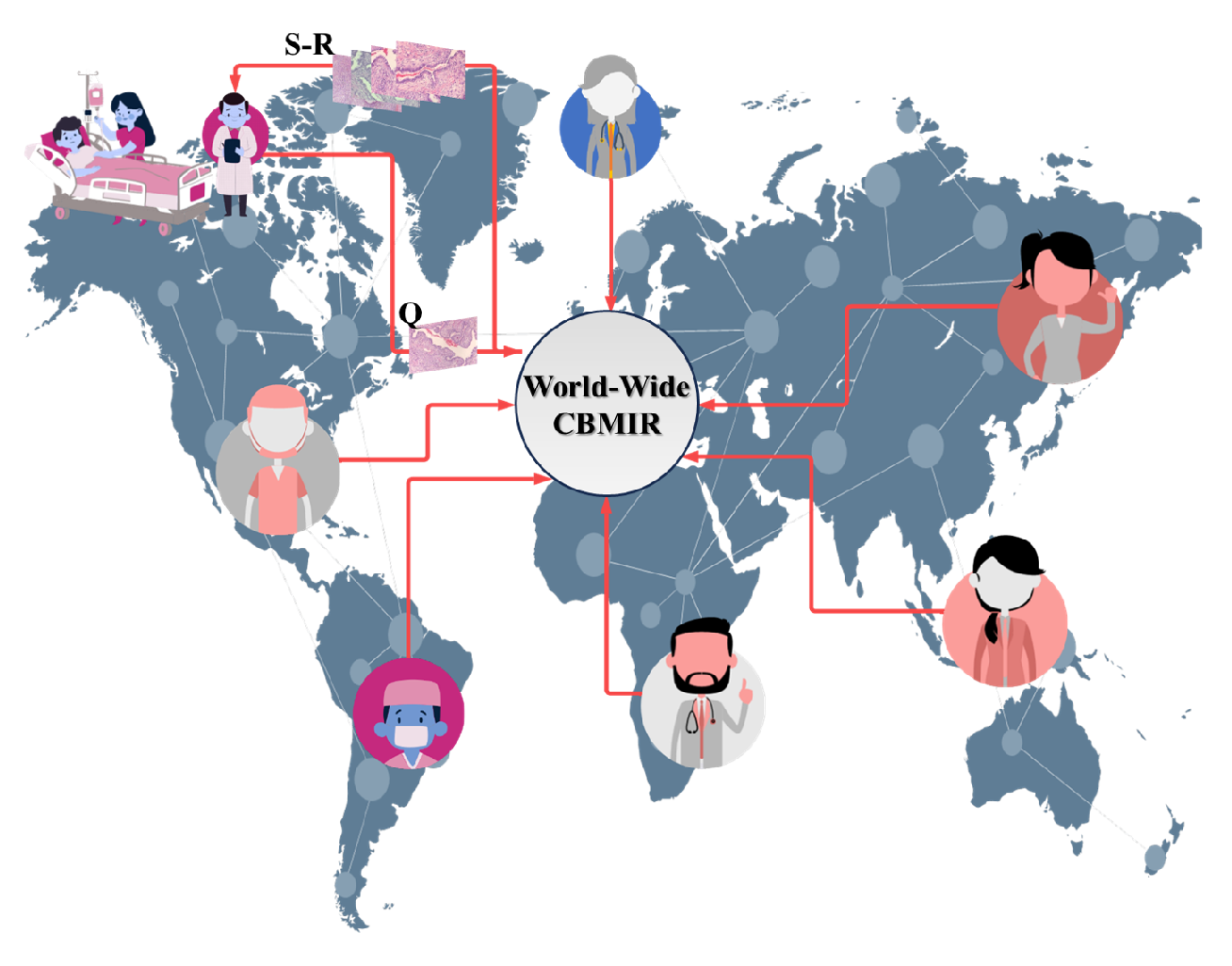
Figure 1. An overview of the use case of a worldwide CBMIR. Pathologists send their query (Q) to the worldwide CBMIR since they need a second opinion to make a more confident decision. Then, the model retrieved top K similar images (S-R), and the pathologists can obtain a second opinion from whole over the world.
One of the advantages of CBMIR from the pathologist’s (user) perspective is that it is not a completely black box for them. CBMIR allows pathologists to find similar patterns among the retrieved images and the queries based on their knowledge. This provides more reliable information than a label for pathologists, which makes CBMIR more beneficial for pathologists than a classification.
An actual context needs a global CBMIR, which demands a generalized data set with a variety of images of different quality, magnification, color, size, etc. The performance of CBMIR relies on a vast amount of data, which is difficult to collect in the medical field due to patient privacy and time costs. In order to create a vast centralized data set, DL experts need to transfer their WSIs. However, these images are gigapixels with high storage sizes. In addition to the challenges of transferring a heavy data set for DL experts, patient privacy policies and other regulatory obstacles on the medical side make it more challenging to create a sufficient data set.
Federated learning (FL) represents a possible solution to tackle this problem by collaboratively training DL models without transferring WSIs [11]. Multiple institutions can safely co-train DL models in digital pathology using FL, achieving cutting-edge performance with privacy assurances [12]. FL brings an opportunity to share the weights for multi-institutional training without sharing patient data and images. However, there are still some privacy risks since the training parameters and model weights are distributed among collaborators [13].
DL models give information that goes beyond the scope of human vision, and FL solves the problem of data sparsity by connecting international hospitals while complying with the data privacy policies, irrespective of the country of origin. This benefit can remedy the health care limitations due to the lack of facilities (staining materials, scanners, etc.) and experience (students, recently graduated pathologists, etc.). Moreover, it can tackle the lack of data sets of labeled WSIs because of data privacy.
2. Content-Based Medical Image Retrieval (CBMIR)
CBMIR has been a subject of extensive research since the advent of large-scale databases nearly two decades ago, as noted by Wang [14]. Several studies have made significant contributions to this field. Tabatabaei [15] achieved an accuracy rate of 84% in CBMIR using the largest patch-annotated data set in prostate cancer. Kalra [16] proposed Yottixel, a method for representing the Cancer Genome Atlas whole-slide images (TCGA WSIs) compactly to facilitate millions of high-accuracy searches with low storage requirements in real time. Conversely, Mehta [17] proposed a CBMIR system for sub-images in high-resolution digital pathology images, utilizing scale-invariant feature extraction. Lowe [18] utilized scale-invariant feature transform (SIFT) to index sub-images and reported an 80% accuracy rate for the top-five retrieved images. Lowe’s experiments were conducted on 50 ImmunohHistoChemistry (IHC) stained pathology images at eight different resolutions. Additionally, Hegde [19] used a manually annotated data set pre-trained on a deep neural network (DNN) to achieve top-five scores for patch-based CBMIR at different magnification levels. The primary focus of recent studies has been on enhancing the performance of CBMIR in different types of cancer; however, there are still several challenges that can impede its effectiveness. These challenges include data privacy, as medical data is confidential and subject to strict privacy regulations, making it arduous to share and access large data sets for model training. FL can alleviate this issue by facilitating distributed model training on local data without compromising privacy. Another challenge is data distribution; as medical data is frequently dispersed across numerous locations, it is difficult to train models on a centralized data set. FL enables the training of models across multiple distributed data sets without aggregating the data in a central location. In addition, medical data sets can be heterogeneous, varying in terms of imaging modalities, quality, and annotation protocols, which can impede the development of robust and accurate models. FL can mitigate this challenge by allowing models to be trained on diverse data sets in different qualities, improving their performance and generalization ability. Furthermore, medical data sets can be large and complex, necessitating significant computational resources to train models. FL can distribute the computational workload across multiple devices and locations, enhancing scalability and reducing training time.
3. Federated Learning (FL)
In recent years, FL has achieved impressive progress that enhances a wide adoption of DL from decentralized data [11][20][21]. FL is a distributed machine learning approach that can effectively handle decentralized data without raw data exchange to train a joint model by aggregating and distributing local training. Many existing algorithms can be adopted to aggregate updates from distributed clients. Typical examples include FederatedAveraging, viz FedAvg [11], and adaptive federated optimization methods [21], e.g., FedAdagrad, FedYogi, and FedAdam. Some popular FL frameworks, such as TensorFlow Federated (TFF) (https://www.tensorflow.org/federated (accessed on 23 September 2022)), PySyft [22], and Flower [23] provide a set of robust tools for building privacy-preserving ML models. In addition, Jupyter-Notebook-based tools, such as [24], also help simplify the FL setup and enable its deployment of a cross-country federated environment in only a few minutes. Daniel Truhn in [25] employed homomorphic encryption to protect the model’s performance while training by encrypting the weight updates before sharing them with the central server. Firas Khader in [26] presented a technique of “learnable synergy”, where the model only chooses pertinent interactions between data modalities and maintains an“internal memory” of key information. Micah J. Sheller [13] investigated how FL among ten institutions is 99% as efficient as that derived using centralized data. One recent work related to content-based image retrieval is introduced in [27], where FLSIR was proposed, and it enables secure image retrieval based on FL and additive secret sharing. Nevertheless, it is not for clinical applications. Although the combination of CBMIR and FL is a relatively new area of research, it has the potential to greatly improve healthcare outcomes. By offering healthcare professionals quick access to accurate and relevant medical image data while maintaining patient privacy, the integration of these techniques can have a significant impact on the field.
References
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S.; et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022, 66, 15–23.
- Zhao, T.; Fu, C.; Tie, M.; Sham, C.W.; Ma, H. RGSB-UNet: Hybrid Deep Learning Framework for Tumour Segmentation in Digital Pathology Images. Bioengineering 2023, 10, 957.
- Rashmi, R.; Prasad, K.; Udupa, C.B.K. Breast histopathological image analysis using image processing techniques for diagnostic purposes: A methodological review. J. Med. Syst. 2022, 46, 7.
- Morelli, P.; Porazzi, E.; Ruspini, M.; Restelli, U.; Banfi, G. Analysis of errors in histology by root cause analysis: A pilot study. J. Prev. Med. Hyg. 2013, 54, 90.
- World Health Organization. Laboratory Quality Standards and Their Implementation; World Health Organization: Geneva, Switzerland, 2011.
- Kim, M.Y.; Choi, N.; Yang, J.H.; Kim, S.; Shin, S.J.; Park, M.H.; Moon, W.; Yoo, Y.B.; Kim, W.H.; Ko, E.Y. Diagnostic accuracy of breast cancer in core needle biopsy using a standardized reporting system. J. Clin. Pathol. 2012, 65, 790–794.
- Baxi, V.; Edwards, R.; Montalto, M.; Saha, S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod. Pathol. 2022, 35, 23–32.
- Fuster, S.; Khoraminia, F.; Kiraz, U.; Kanwal, N.; Kvikstad, V.; Eftestøl, T.; Zuiverloon, T.C.; Janssen, E.A.; Engan, K. Invasive cancerous area detection in Non-Muscle invasive bladder cancer whole slide images. In Proceedings of the 2022 IEEE 14th Image, Video, and Multidimensional Signal Processing Workshop (IVMSP), Nafplio, Greece, 26–29 June 2022; pp. 1–5.
- Zheng, Y.; Jiang, Z.; Zhang, H.; Xie, F.; Ma, Y.; Shi, H.; Zhao, Y. Size-scalable content-based histopathological image retrieval from database that consists of WSIs. IEEE J. Biomed. Health Inform. 2017, 22, 1278–1287.
- Baâzaoui, A.; Abderrahim, M.; Barhoumi, W. Dynamic distance learning for joint assessment of visual and semantic similarities within the framework of medical image retrieval. Comput. Biol. Med. 2020, 122, 103833.
- McMahan, B.; Moore, E.; Ramage, D.; Hampson, S.; y Arcas, B.A. Communication-efficient learning of deep networks from decentralized data. In Proceedings of the Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 20–22 April 2017; pp. 1273–1282.
- Lu, M.Y.; Chen, R.J.; Kong, D.; Lipkova, J.; Singh, R.; Williamson, D.F.; Chen, T.Y.; Mahmood, F. Federated learning for computational pathology on gigapixel whole slide images. Med. Image Anal. 2022, 76, 102298.
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated learning in medicine: Facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 2020, 10, 12598.
- Wang, W.; Jiao, P.; Liu, H.; Ma, X.; Shang, Z. Two-stage content based image retrieval using sparse representation and feature fusion. Multimed. Tools Appl. 2022, 81, 16621–16644.
- Tabatabaei, Z.; Colomer, A.; Engan, K.; Oliver, J.; Naranjo, V. Residual block Convolutional Auto Encoder in Content-Based Medical Image Retrieval. In Proceedings of the 2022 IEEE 14th Image, Video, and Multidimensional Signal Processing Workshop (IVMSP), Nafplio, Greece, 26–29 June 2022; pp. 1–5.
- Kalra, S.; Tizhoosh, H.R.; Choi, C.; Shah, S.; Diamandis, P.; Campbell, C.J.; Pantanowitz, L. Yottixel–an image search engine for large archives of histopathology whole slide images. Med. Image Anal. 2020, 65, 101757.
- Mehta, N.; Alomari, R.S.; Chaudhary, V. Content based sub-image retrieval system for high resolution pathology images using salient interest points. In Proceedings of the 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Minneapolis, MN, USA, 3–6 September 2009; pp. 3719–3722.
- Lowe, D.G. Object recognition from local scale-invariant features. In Proceedings of the Seventh IEEE International Conference on Computer Vision, Kerkyra, Greece, 20–27 September 1999; Volume 2, pp. 1150–1157.
- Hegde, N.; Hipp, J.D.; Liu, Y.; Emmert-Buck, M.; Reif, E.; Smilkov, D.; Terry, M.; Cai, C.J.; Amin, M.B.; Mermel, C.H.; et al. Similar image search for histopathology: SMILY. NPJ Digit. Med. 2019, 2, 56.
- Bonawitz, K.; Eichner, H.; Grieskamp, W.; Huba, D.; Ingerman, A.; Ivanov, V.; Kiddon, C.; Konečnỳ, J.; Mazzocchi, S.; McMahan, B.; et al. Towards federated learning at scale: System design. Proc. Mach. Learn. Syst. 2019, 1, 374–388.
- Reddi, S.; Charles, Z.; Zaheer, M.; Garrett, Z.; Rush, K.; Konečnỳ, J.; Kumar, S.; McMahan, H.B. Adaptive federated optimization. arXiv 2020, arXiv:2003.00295.
- Ziller, A.; Trask, A.; Lopardo, A.; Szymkow, B.; Wagner, B.; Bluemke, E.; Nounahon, J.M.; Passerat-Palmbach, J.; Prakash, K.; Rose, N.; et al. Pysyft: A library for easy federated learning. In Federated Learning Systems: Towards Next-Generation AI; Springer: Cham, Switzerland, 2021; pp. 111–139.
- Beutel, D.J.; Topal, T.; Mathur, A.; Qiu, X.; Parcollet, T.; de Gusmão, P.P.; Lane, N.D. Flower: A friendly federated learning research framework. arXiv 2020, arXiv:2007.14390.
- Launet, L.; Wang, Y.; Colomer, A.; Igual, J.; Pulgarín-Ospina, C.; Koulouzis, S.; Bianchi, R.; Mosquera-Zamudio, A.; Monteagudo, C.; Naranjo, V.; et al. Federating Medical Deep Learning Models from Private Jupyter Notebooks to Distributed Institutions. Appl. Sci. 2023, 13, 919.
- Truhn, D.; Arasteh, S.T.; Saldanha, O.L.; Müller-Franzes, G.; Khader, F.; Quirke, P.; West, N.P.; Gray, R.; Hutchins, G.G.; James, J.A.; et al. Encrypted federated learning for secure decentralized collaboration in cancer image analysis. medRxiv 2022.
- Khader, F.; Mueller-Franzes, G.; Wang, T.; Han, T.; Arasteh, S.T.; Haarburger, C.; Stegmaier, J.; Bressem, K.; Kuhl, C.; Nebelung, S.; et al. Medical Diagnosis with Large Scale Multimodal Transformers–Leveraging Diverse Data for More Accurate Diagnosis. arXiv 2022, arXiv:2212.09162.
- Zhang, L.; Xia, R.; Tian, W.; Cheng, Z.; Yan, Z.; Tang, P. FLSIR: Secure Image Retrieval Based on Federated Learning and Additive Secret Sharing. IEEE Access 2022, 10, 64028–64042.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
629
Revisions:
2 times
(View History)
Update Date:
17 Oct 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




