Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | WenWei Li | -- | 2114 | 2023-09-26 10:41:11 | | | |
| 2 | Lindsay Dong | Meta information modification | 2114 | 2023-09-28 02:35:04 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Li, W.; Zhou, Z.; Wang, D. Advances in the Biosynthesis of L-Cysteine. Encyclopedia. Available online: https://encyclopedia.pub/entry/49648 (accessed on 28 February 2026).
Li W, Zhou Z, Wang D. Advances in the Biosynthesis of L-Cysteine. Encyclopedia. Available at: https://encyclopedia.pub/entry/49648. Accessed February 28, 2026.
Li, Wenwei, Zhen Zhou, Dan Wang. "Advances in the Biosynthesis of L-Cysteine" Encyclopedia, https://encyclopedia.pub/entry/49648 (accessed February 28, 2026).
Li, W., Zhou, Z., & Wang, D. (2023, September 26). Advances in the Biosynthesis of L-Cysteine. In Encyclopedia. https://encyclopedia.pub/entry/49648
Li, Wenwei, et al. "Advances in the Biosynthesis of L-Cysteine." Encyclopedia. Web. 26 September, 2023.
Copy Citation
L-Cysteine is a widely used unique sulfur-containing amino acid with wide application in the food, pharmaceutical, and agricultural industries.
L-cysteine
Escherichia coli
metabolic engineering
synthetic biology
1. Introduction
L-cysteine is a sulfur-containing amino acid that plays an important role in the folding of proteins, has a high redox activity in cellular metabolism, is a catalytic residue for a variety of enzymes, and is a sulfur donor compound that is required for the synthesis of Fe/S clusters, biotin, coenzyme A, and thiamine [1][2][3]. In addition to its roles in cellular metabolism, L-cysteine plays a variety of roles in metal binding, catalytic activity, and redox and has a vast array of industrial applications in the production of food, cosmetics, pharmaceuticals, and animal feed [4][5][6].
Chemical hydrolysis of proteins, which are typically extracted from the keratin of animal hair such as feathers, pig hair, etc., is the traditional method of industrial production of L-cysteine [7]. However, this method not only consumes a large amount of hydrochloric acid, but it also causes an unpleasant odor and wastewater treatment problems, which have a significant impact on the environment [7]. To avoid the environmental hazards of this method, scientists have explored biotechnological approaches to synthesize L-cysteine as an alternative to chemical hydrolysis. Fermentation and enzymatic biotransformation are the two most prominent biotechnological methods [8][9]. However, due to the presence of generated L-cysteine products that inhibit the activity of the enzymes, the enzyme bioconversion method presents difficulty in solving the problems of low yield and high cost.
Although fermentation offers a number of advantages, the design and construction of efficient microbial cell factories for fermentative production of L-cysteine remains challenging due to the high toxicity of L-cysteine and the complex regulation of its synthetic pathway [10]. The efficient production of L-cysteine on an industrial scale has not yet been achieved, which is a major challenge for the industrialization of L-cysteine [11][12]. Numerous microorganisms, including bacteria such as E. coli, C. glutamicum, and Pantoea ananatis, have been engineered to produce L-cysteine due to the rapid development of systems metabolic engineering and synthetic biology [13][14][15][16][17]. In comparison to other bacteria, E. coli has a rapid growth rate and more developed genetic engineering techniques, whereas C. glutamicum is a non-pathogenic, industrial microorganism with developed fermentation technology that is extensively employed in food processing and other industries [13][18]. Therefore, E. coli and C. glutamicum are the two most studied chassis cells that directly produce L-cysteine from glucose [19][20][21].
2. Advances in the Biosynthesis of L-Cysteine
2.1. Enzyme Biotransformation—Asymmetrical Hydrolysis of DL-2-amino-Δ2-thiazoline-4- Carboxylic Acid
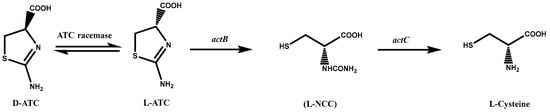
Since L-cysteine is traditionally obtained by hydrolyzing animal hair, the extraction of 1 kg of L-cysteine requires approximately 10 kg of animal hair and 2.7 kg of hydrochloric acid, a process that not only has a low yield but also produces foul odors and wastewater, causing severe environmental damage [22]. The transformation method uses Pseudomonas to enzymatically convert DL-2-amino-Δ2-thiazoline-4-carboxylic acid (DL-ATC) to L-cysteine. This method of converting DL-ATC to cysteine involves three enzymes: ATC racemase, L-ATC hydrolase, and S-carbamoyl-L-cysteine hydrolase [23][24]. The complete procedure consists of three stages (Figure 1): (i) conversion of D-ATC to L-ATC by ATC racemase, (ii) ring-opening of L-ATC by L-ATC hydrolase to produce N-carbamoyl-L-cysteine (L-NCC), and (iii) final hydrolysis of L-NCC to L-cysteine by S-carbamoyl-L-cysteine hydrolase [25].

Figure 1. A metabolic pathway of DL-2-amino-Δ2-thiazoline-4-carboxylic acid (DL-ATC) to L-cysteine via N-carbamyl-L-cysteine (L-NCC) in Pseudomonas species. atcB gene encoding L-ATC acid hydrolase; atcC gene encoding L-NCC amidohydrolase.
Both L-ATC hydrolase (atcB) and S-carbamoyl-L-cysteine hydrolase (atcC) genes originated from Pseudomonas sp. strain BS. After sequencing by Japanese scientists, the amino acid sequence of the atcC gene product was found to be highly homologous to L-N-carbamoylase from other bacteria, but the amino acid sequence of the atcB gene was novel [8]. AtcB was initially identified as a gene encoding an enzyme that catalyzes the thiazoline ring-opening reaction and does not share a high degree of homology with previously described enzymes [26].
Enzymatic bioconversion is to some extent environmentally friendly and has lower energy consumption than hydrolysis of animal hair. However, the high toxicity of L-cysteine inhibits enzyme activity, leading to low efficiency and relatively high cost [8][9].
2.2. Biological Fermentation Methods
2.2.1. L-Cysteine Biosynthesis in E. coli
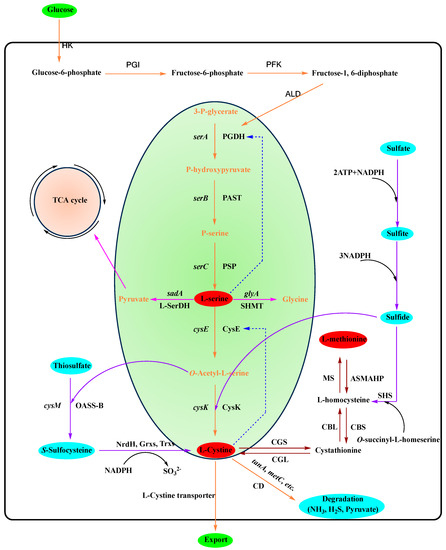
It is well known that in most microorganisms and plants, L-serine is the precursor substance for the synthesis of L-cysteine. The biosynthetic pathway of L-cysteine has been widely reported after many years of research [4][27][28]. In gut bacteria, L-serine is synthesized via a three-step pathway from the glycolytic intermediate 3-phosphoglycerate, and L-cysteine is synthesized via a two-step pathway from L-serine [28]. Firstly, the glycolytic intermediate 3-phosphoglycerate is converted to L-serine by a three-step reaction catalyzed by 3-phosphoglycerate dehydrogenase (PGDH), phosphoserine aminotransferase (PAST), and phosphoserine phosphatase (PSP) (Figure 2) [27].

Figure 2. The metabolic pathway of L-cysteine in E. coli and C. glutamicum. Brown arrows refer to metabolic pathways in P. aeruginosa. The solid purple line indicates the conversion of L-serine to other productsDashed lines represent feedback inhibition. The solid orange line indicates the major metabolic pathway of L-cysteine. The solid purple line indicates the source of the sulfide. Italicized fonts on the same arrow line indicate genes encoding corresponding enzymes. HK, hexokinase; PGI, phosphoglucose isomerase; PFK, phosphofructokinase; ALD, aldolase; PGDH, 3-phosphoglycerate dehydrogenase; PAST, phosphoserine aminotransferase; PSP, phosphoserine phosphatase; CysE, serine O-acetyltransferase; CysK, cysteine synthase; L-SerDH, L-serine dehydratase; SHMT, serine hydroxymethyl transferase; CD, L-cysteine desulfhydrases; OASS-B, O-acetyl-L-serine sulfhydrylase-A; NrdH and Grxs, glutaredoxins; Trxs, thioredoxins; CGS, cystathionine γ-synthase; CBL, cystathionine β-lyase; MS, methionine synthase; SHS, O-succinyl-L-homoserine sulfhydrylase; ASMAHP, S-adenosylmethionine synthase-methyltransferases-S-adenosylhomocysteine hydrolase pathway; CBS, cystathionine β-synthase; CGL, cystathionine γ-lyase.
Scientists have conducted studies to address the aforementioned causes of L-cysteine biosynthesis blockage. Table 1 summarizes the research progress on the fermentation synthesis of L-cysteine by different engineering strains.
Table 1. Progress of fermentation synthesis of L-cysteine by different engineered strains.
| Bacterial Strain | Metabolic Strategy | L-Cysteine Production (g/L) | Productivity (g/(L·h)) | References |
|---|---|---|---|---|
| E. coli JM240 | Enhancing biosynthesis | 0.03 | / | [29] |
| E. coli JM39 | Enhancing biosynthesis | 0.20 | 0.003 | [30] |
| E. coli W3110 | Enhancing excretion | 0.07 | 0.003 | [31] |
| E. coli W3110 | Enhancing excretion | 0.15 | 0.007 | [32] |
| E. coli JM39 | Enhancing biosynthesis and weakening degradation | 0.60 | 0.013 | [33] |
| E. coli MG1655 | Enhancing biosynthesis and excretion and weakening degradation | 1.20 | 0.025 | [34] |
| E. coli BW25113 | Enhancing biosynthesis and excretion | 1.23 | 0.026 | [35] |
| E. coli BW25113 | Enhancing biosynthesis and excretion/weakening degradation | 1.72 | 0.024 | [20] |
| E. coli JM109 | Enhancing the sulfur conversion rate | 7.50 | 0.341 | [14] |
| E. coli BW25113 | Enhancing biosynthesis and thiosulfate assimilation and weakening degradation | 8.34 | 0.321 | [10] |
| E. coli W3110 | Balancing carbon and sulfur module conversion rate | 11.94 | 0.254 | [36] |
| C. glutamicum IR33 | Enhancing biosynthesis | 0.29 | 0.004 | [37] |
| C. glutamicum ATCC13032; C. glutamicum ATCC21586 |
Enhanced sulfur metabolism in biosynthesis | 0.06 | 0.004 | [27] |
| C. glutamicum NBRC12168 | Enhancing biosynthesis and weakening degradation | 0.20 | 0.017 | [16] |
| C. glutamicum CYS | Enhancing biosynthesis and excretion | 0.28 | 0.014 | [19] |
| C. glutamicum ATCC13032 | Enhancing precursor accumulation and weakening degradation | 0.95 | 0.026 | [15] |
| C. glutamicum Cys -10 |
Enhancing biosynthesis, excretion, and sulfur metabolism and weakening degradation | 5.92 | 0.082 | [38] |
| Pantoea ananatis | Weakening degradation and educing efflux | 2.20 | 0.079 | [17] |
2.2.2. L-Cysteine Biosynthesis in C. glutamicum
The metabolic pathway of L-cysteine in C. glutamicum is generally the same as that of E. coli, and L-serine is also synthesized from the glycolytic intermediate 3-phosphoglyceric acid via a three-step pathway, followed by further conversion to L-cysteine via CysE and CysK [39]. However, wild-type C. glutamicum is subjected to a feedback mechanism that produces almost no L-cysteine. In order to allow C. glutamicum to produce L-cysteine, the cysE gene (the gene is insensitive to feedback inhibition by L-cysteine) encoding an alteration of CysE in the E. coli Met256Ile mutant was introduced into C. glutamicum, which produces approximately 0.29 g/L of L-cysteine [37]. CysR is a transcriptional regulator that regulates sulfur metabolism in L-cysteine biosynthesis, and overexpression of the cysR gene in C. glutamicum resulted in a 2.7-fold higher intracellular sulfide concentration than that of the control strain (empty pMT-tac vector), and overexpression of the cysE, cysK, and cysR genes in C. glutamicum resulted in a 3-fold higher L-cysteine production than that of the control [27].
2.2.3. L-Cysteine Biosynthesis in Other Bacteria
It is reported that in addition to E. coli and C. glutamicum, several other microbial bacteria have been reported to produce L-cysteine, such as Saccharomyces cerevisiae, Lactococcus lactis, Mycobacterium tuberculosis, and Pseudomonas species, which can synthesize L-cysteine from L-methionine by the reverse transsulfuration pathway [40][41][42][43][44]. However, it was later demonstrated that L-cysteine synthesis in Saccharomyces cerevisiae is exclusively by L-cystathionine β-synthase (CBS; EC 4.2.1.22) and L-cystathionine γ-lyase (CGL; EC 4.4.1.1) via cystathionine [45][46].
3. Metabolic Engineering Strategies for L-Cysteine Biosynthetic Pathway
3.1. Enhanced Accumulation of Precursor L-Serine
The precursor of L-cysteine is L-serine, and enhanced accumulation of L-serine is essential to increase the yield of the target amino acid L-cysteine. Firstly, the metabolic flux from 3-phosphoglycerate to L-serine can be improved, or feedback inhibition caused by high L-serine titers can be reduced, and enzymes in the L-serine biosynthesis pathway have to be regulated to be overexpressed or genetically altered [47]. Secondly, it is important to prevent the degradation of L-serine, which is not only a precursor of L-cysteine but also a precursor of glycine, L-alanine, and L-valine and is even necessary for protein synthesis, phospholipid synthesis, and C1 unit production [48][49]. Therefore, the enzymes of the relevant degradation pathways must be inhibited, or the corresponding genes must be knocked out to ensure that L-serine is converted to L-cysteine as much as possible [50][51][52].
3.2. Increase the Accumulation of Sulfides
Sulfur in L-cysteine, found in plants and bacteria, is primarily derived from sulfate. The sulfur assimilation processes of L-cysteine in E. coli are categorized into two main pathways: sulfate assimilation and thiosulfate assimilation pathways. In the sulphate assimilation pathway, O-acetyl-L-serine sulfhydrylase A catalyzes the conversion of OAS and sulfide (S2−) into L-cysteine [14]. In the thiosulfate assimilation pathway, the entry of inorganic sulfur into the cell is achieved through the thiosulfate ABC transporter proteins, which are encoded by the genes cysU, cysW, cysA, and sbp. Thiosulfate is not only less energy intensive than sulfate but also provides a more efficient source of sulfur, making it the current optimal choice [35][53][54]. As thiosulfate is transported into the cytoplasm, it reacts with OAS via the catalytic action of O-acetyl-L-serine sulfhydrylase B, with the consequent formation of S-sulfo-L-cysteine (SSC). SSC is ultimately converted to L-cysteine by the enzymes NrdH and GrxA [55]. Thus, optimizing the thiosulfate pathway can enhance sulfide accumulation, thereby enhancing L-cysteine production.
3.3. Decrease the Degradation of L-Cysteine
L-cysteine degradation in E. coli is primarily catalyzed by the enzyme CD, which catalyzes L-cysteamine degradation to pyruvate, ammonia, and sulfide [50]. Currently, it has been determined that the degradation activity of the enzyme CD is encoded by the aceD gene, and disruption of the aceD gene, which encodes the degradation activity of the CD enzyme, has been reported to slow down the degradation of L-cysteine [16][50]. However, the concentration of L-cysteine continues to decrease during the stationary growth phase [16].
3.4. Enhance the Ability of Cells to Output L-Cysteine
Advances in synthetic biology and systems metabolic engineering have provided various tools and techniques for engineering microbial strains for the production of bio-based chemicals [56]. Among these methods, efficient output of target products is a very promising method that can improve the robustness and efficiency of microbial cell factories, providing significant assistance in improving substrate absorption, overcoming metabolic inhibition, protecting cells from toxic compounds, and providing assistance for efficient fermentation production of target products [57][58].
4. Challenges and New Ideas in the Biological Production of L-Cysteine
4.1. In Vitro Metabolic Pathways
In vitro metabolic engineering is an alternative production technology for fermentation that utilizes a cascade reaction composed of purified/semi purified enzymes for chemical production [59][60]. During production, there are no living cells in in vitro metabolic engineering, so there is no impact of toxic products on cell activity [61]. Recently, it was reported that L-cysteine was produced by in vitro metabolic engineering and that 28.2 mM L-cysteine was produced from 20 mM glucose with a molar yield of 70.5%, surpassing the greatest yield of L-cysteine produced by fermentation at present [62]. In this reaction, the genes for the enzymes needed to synthesize L-cysteine in vitro are put together in a single expression vector and co-expressed in a single strain.
4.2. Explore New Raw Materials
Glycerol is one of the main by-products in biodiesel production, and according to statistics, every 10 kg of biodiesel produces approximately 1 kg of crude glycerol by-products, so utilizing glycerol would be a great asset [63][64]. With continuous research, the conversion of glycerol into high-value-added products has become more competitive [65][66]. The production of L-cysteine from E. coli using glycerol instead of glucose as substrate has been recently reported to have been achieved [21]. The metabolic pathway from glycerol to L-cysteine is shorter, and the carbon atom economy is more favorable compared to glucose [67][68].
4.3. Utilization of New Technologies
The desire for efficient microbial cell factories requires constant regulation of metabolism and modulation of gene expression, which is often time-consuming [69]. The emergence of new technologies, such as CRISPR-Cpf1 gene editing and high-throughput screening (HTS), has provided a significant boost to metabolic engineering as a result of the development of synthetic biology [70][71]. In recent years, an efficient CRISPR-Cpf1 genome-editing system has been established in C. glutamicum, which holds promise for improving L-cysteine production [72][73][74].
References
- Cicchill, R.M.; Baker, M.A.; Schnitze, E.J.; Newman, E.B.; Krebs, C.; Booker, S.J. E. coli L-Serine Deaminase Requires a Cluster in Catalysis. J. Biol. Chem. 2004, 279, 32418–32425.
- Ohnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005, 74, 247–281.
- Heieck, K.; Arnold, N.D.; Brück, T.B. Metabolic stress constrains microbial L-cysteine production in E. coli by accelerating transposition through mobile genetic elements. Microb. Cell Fact. 2023, 22, 10.
- Takagi, H.; Ohtsu, I. L-cysteine metabolism and fermentation in microorganisms. Adv. Biochem. Eng. Biotechnol. 2016, 159, 129–151.
- Colovic, M.B.; Vasic, V.M.; Djuric, D.M.; Krstic, D.Z. Sulphur-containing Amino Acids: Protective Role Against Free Radicals and Heavy Metals. Curr. Med. Chem. 2018, 25, 324–335.
- Wendisch, V.F. Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 2020, 58, 17–34.
- Ismail, N.R.; Hashim, Y.Z.; Jamal, P.; Othman, R.; Salleh, H.M. Production of Cysteine: Approaches, Challenges and Potential Solution. Int. J. Biotechnol. Wellness Indus. 2014, 3, 95–101.
- Shiba, T.; Takeda, K.; Yajima, M.; Tadano, M. Genes from Pseudomonas sp. Strain BS Involved in the Conversion of L-2-Amino-Δ2-Thiazolin-4-Carbonic Acid to L-Cysteine. Appl. Environ. Microbiol. 2002, 68, 2179–2187.
- Nam, K.H.; Ryu, O.H.; Park, J.; Shin, C.S. Effects of anoxic conditions on the enzymatic conversion of D, L-2-amino-thiazoline-4-carboxylic acid to L-cystine. Acta Biotechnol. 1997, 17, 185–193.
- Liu, H.; Wang, Y.; Hou, Y.; Li, Z. Fitness of chassis cells and metabolic pathways for L-cysteine overproduction in E. coli. J. Agric. Food Chem. 2020, 68, 14928–14937.
- Yin, J.; Ren, W.; Yang, G.; Duan, J.; Huang, X.; Fang, R.; Li, C.; Li, T.; Yin, Y.; Hou, Y.Q.; et al. L-Cysteine metabolism and its nutritional implications. Mol. Nutr. Food Res. 2016, 60, 134–146.
- Liu, G.; Ding, C.; Ju, Y.; Ma, Z.; Wei, L.; Liu, J.; Liu, Q.; Xu, N. Directed evolution of an EamB transporter for improved L-cysteine tolerance and production in E. coli. FEMS Microbiol. Lett. 2021, 368, fnac008.
- Liu, H.; Fang, G.; Wu, H.; Li, Z.; Ye, Q. L-Cysteine production in E. coli based on rational metabolic engineering and modular strategy. Biotechnol. J. 2018, 13, e1700695.
- Liu, H.; Hou, Y.; Wang, Y.; Li, Z. Enhancement of sulfur conversion rate in the production of L-cysteine by engineered E. coli. J. Agric. Food Chem. 2020, 68, 250–257.
- Wei, L.; Wang, H.; Xu, N.; Zhou, W.; Ju, J.; Liu, J.; Ma, Y. Metabolic engineering of C. glutamicum for L-cysteine production. Appl. Microbiol. Biotechnol. 2019, 103, 1325–1338.
- Kondoh, M.; Hirasawa, T. L-Cysteine production by metabolically engineered C. glutamicum. Appl. Microbiol. Biotechnol. 2019, 103, 2609–2619.
- Takumi, K.; Ziyatdinov, M.K.; Samsonov, V.; Nonaka, G. Fermentative production of cysteine by Pantoea ananatis. Appl. Environ. Microbiol. 2017, 83, e02502-16.
- Yang, J.; Yang, S. Comparative analysis of C. glutamicum genomes: A new perspective for the industrial production of amino acids. BMC Genom. 2017, 1, 940.
- Kishino, K.; Kondoh, M.; Hirasawa, T. Enhanced L-cysteine production by overexpressing potential L-cysteine exporter genes in an L-cysteine-producing recombinant strain of C. glutamicum. Biosci. Biotechnol. Biochem. 2019, 83, 2390–2393.
- Kawano, Y.; Ohtsu, I.; Takumi, K.; Tamakoshi, A.; Nonaka, G.; Funahashi, E.; Ihara, M.; Takagi, H. Enhancement of L-cysteine production by disruption of yciW in E. coli. J. Biosci. Bioeng. 2015, 119, 176–179.
- Zhang, X.; Sun, Z.; Bian, J.; Gao, Y.; Zhang, D.; Xu, G.; Zhang, X.; Li, H.; Shi, J.; Xu, Z. Rational Metabolic Engineering Combined with Biosensor-Mediated Adaptive Laboratory Evolution for L-Cysteine Overproduction from Glycerol in E. coli. Fermentation 2022, 8, 299.
- Renneberg, R. High grade cysteine no longer has to be extraxted from hair. In Biotechnology for Beginners; Demain, A.L., Ed.; Academic Press: Amsterdam, The Netherlands, 2008; p. 106.
- Sano, K.; Mitsugi, K. Enzymatic production of L-cysteine from DL-2-amino-Δ2-thiazoline-4-carboxylic acid by Pseudomonas thiazolinophium: Optimal conditions for the enzymeformation and enzymatic reaction. Agric. Biol. Chem. 1978, 42, 2315–2321.
- Sano, K.; Eguchi, C.; Yasuda, N.; Mitsugi, K. Metabolic pathway for L-cysteine formation from DL-2-amino-Δ2-thiazoline-4-carboxylic acid by Pseudomonas. Agric. Biol. Chem. 1979, 43, 2373–2374.
- Tamura, T.; Nishino, M.; Ohmachi, T.; Asada, Y. N-Carbamoyl-L-Cysteine as an Intermediate in the Bioconversion from D, L-2-Amino-Δ2-Thiazoline-4-Carboxylic Acid to L-Cysteine by Pseudomonas sp. ON-4a. Biosci. Biotechnol. Biochem. 1998, 62, 2226–2229.
- Ohmachi, T.; Nishino, M.; Kawata, M.; Edo, N.; Funaki, H.; Narita, M.; Mori, K.; Tamura, Y.; Asada, Y. Identification, cloning, and sequencing of the genes involved in the conversion of D, L-2-amino-Δ2-thiazoline-4-carboxylic acid to L-cysteine in Pseudomonas sp. strain ON-4a. Biosci. Biotechnol. Biochem. 2002, 66, 1097–1104.
- Joo, Y.C.; Hyeon, J.E.; Han, S.O. Metabolic Design of C. glutamicum for Production of L-Cysteine with Consideration of Sulfur-Supplemented Animal Feed. J. Agric. Food Chem. 2017, 65, 4698–4707.
- Wada, M.; Takagi, H. Metabolic pathways and biotechnological production of L-cysteine. Appl. Microbiol. Biot. 2006, 73, 48–54.
- Denk, D.; Böck, A. L-Cysteine Biosynthesis in E. coli: Nucleotide Sequence and Expression of the Serine Acetyltransferase (cysE) Gene from the Wild-type and a Cysteine-excreting Mutant. J. Gen. Microbiol. 1987, 133, 515–525.
- Nakamori, S.; Kobayashi, S.I.; Kobayashi, C.; Takagi, H. Overproduction of l-Cysteine and L-Cystine by E. coli Strains with a Genetically Altered Serine Acetyltransferase. Appl. Environ. Microbiol. 1998, 64, 1607–1611.
- Daßler, T.; Maier, T.; Winterhalter, C.; Böck, A. Identification of a major facilitator protein from E. coli involved in efflux of metabolites of the cysteine pathway. Mol. Microbiol. 2000, 36, 1101–1112.
- Franke, I.; Resch, A.; Daßler, T.; Maier, T.; Böck, A. YfiK from E. coli Promotes Export of O-Acetylserine and Cysteine. J. Bacteriol. 2003, 185, 1161–1166.
- Awano, N.; Wada, M.; Kohdoh, A.; Oikawa, T.; Takagi, H.; Nakamori, S. Effect of cysteine desulfhydrase gene disruption on L-cysteine overproduction in E. coli. Appl. Microbiol. Biotechnol. 2003, 62, 239–243.
- Wiriyathanawudhiwong, N.; Ohtsu, I.; Li, Z.D.; Mori, H.; Takagi, H. The outer membrane TolC is involved in cysteine tolerance and overproduction in E. coli. Appl. Microbiol. Biotechnol. 2009, 81, 903–913.
- Nakatani, T.; Ohtsu, I.; Nonaka, G.; Wiriyathanawudhiwong, N.; Morigasaki, S.; Takagi, H. Enhancement of thioredoxin/glutaredoxin-mediated L-cysteine synthesis from S-sulfocysteine increases L-cysteine production in E. coli. Microb. Cell Fact. 2012, 11, 62.
- Zhang, B.; Chen, K.; Yang, H.; Wu, Z.; Liu, Z.; Zheng, Y. Construction of an L-cysteine hyper-producing strain of E. coli based on a balanced carbon and sulfur module strategy. Chin. J. Biotechnol. 2022, 38, 4567–4586.
- Ada, M.; Awano, N.; Haisa, K.; Takagi, H.; Nakamori, S. Purification, characterization and identification of cysteine desulfhydrase of C. glutamicum, and its relationship to cysteine. FEMS Microbiol. Lett. 2002, 217, 103–107.
- Du, H.M.; Qiao, J.F.; Qi, Y.T.; Li, L.C.; Xu, N.; Shao, L.; Wei, L.; Liu, J. Reprogramming the sulfur recycling network to improve L-cysteine production in C. glutamicum. Green Chem. 2023, 25, 3152–3165.
- Peter-Wendisch, P.; Stolz, M.; Etterich, H.; Kennerknecht, N.; Sahm, H.; Eggeling, L. Metabolic engineering of C. glutamicum for L-serine production. Appl. Environ. Microbiol. 2005, 71, 7139–7144.
- Sperandio, B.; Polard, P.; Ehrlich, D.S.; Renault, P.; Guédon, E. Sulfur Amino Acid Metabolism and Its Control in Lactococcus lactis IL1403. J. Bacteriol. 2005, 187, 3762–3778.
- Vermeji, P.; Kertesz, M.A. Pathways of Assimilative Sulfur Metabolism in Pseudomonas putida. J. Bacteriol. 1999, 181, 5833–5837.
- Wheeler, P.R.; Coldham, N.G.; Keating, L.; Gordon, S.V.; Wooff, E.E.; Parish, T.; Hewinson, R.G. Functional Demonstration of Reverse Transsulfuration in the Mycobacterium tuberculosis Complex Reveals That Methionine Is the Preferred Sulfur Source for Pathogenic Mycobacteria. J. Biol. Chem. 2005, 280, 8069–8078.
- Haitani, Y.; Awano, N.; Yamazaki, M.; Wada, M.; Nakamori, S.; Takagi, H. Functional analysis of L-serine O-acetyltransferase from C. glutamicum. FEMS Microbiol. Lett. 2006, 255, 156–163.
- Huai, L.; Chen, N.; Yang, W.; Bai, G. Metabolic control analysis of L-cysteine producing strain TS1138 of Pseudomonas sp. Biochemistry (Moscow) 2009, 74, 288–292.
- Ono, B.; Hazu, T.; Yoshida, S.; Kawato, T.; Shinoda, S.; Brzvwczy, J.; Paszewski, A. Cysteine biosynthesis in Saccharomyces cerevisiae: A new outlook on pathway and regulation. Yeast 1999, 15, 1365–1375.
- Takagi, H.; Yoshioka, K.; Awano, N.; Nakamori, S.; Ono, B. Role of Saccharomyces cerevisiae serine O-acetyltransferase in cysteine biosynthesis. FEMS Microbiol. Lett. 2003, 218, 291–297.
- Zhang, X.; Xu, G.; Shi, J.; Koffas, M.; Xu, Z. Microbial Production of L-Serine from Renewable Feedstocks. Trends Biotechnol. 2018, 36, 700–712.
- Zhu, Q.; Zhang, X.; Luo, Y.; Guo, W.; Xu, G.; Shi, J.; Xu, Z. L-Serine overproduction with minimization of byproduct synthesis by engineered C. glutamicum. Appl. Microbiol. Biotechnol. 2015, 99, 1665–1673.
- Xu, G.; Zhang, X.; Xiao, W.; Shi, J.; Xu, Z. Production of L-serine and its derivative L-cysteine from renewable feedstocks using C. glutamicum: Advances and perspectives. Crit. Rev. Biotechnol. 2023.
- Netzer, R.; Peters-Wendisch, P.; Eggeling, L.; Sahm, H. Cometabolism of a Nongrowth Substrate: L-Serine Utilization by C. glutamicum. Appl. Environ. Microbiol. 2004, 70, 7148–7155.
- Wieschalka, S.; Blombach, B.; Eikmanns, B.J. Engineering C. glutamicum for the production of pyruvate. Appl. Microbiol. Biotechnol. 2012, 94, 449–459.
- Guo, W.; Chen, Z.; Zhang, X.; Xu, G.; Zhang, X.; Shi, J.; Xu, Z. A novel aceE mutation leading to a better growth profile and a higher L-serine production in a high-yield L-serine-producing C. glutamicum strain. J. Ind. Microbiol. Biotechnol. 2016, 43, 1293–1301.
- Kawano, Y.; Onishi, F.; Shiroyama, M.; Miura, M.; Tanaka, N.; Oshiro, S.; Nonaka, G.; Nakanishi, T.; Ohtsu, I. Improved fermentative L-cysteine overproduction by enhancing a newly identified thiosulfate assimilation pathway in E. coli. Appl. Microbiol. Biotechnol. 2017, 101, 6879–6889.
- Yamazaki, S.; Takei, K.; Nonaka, G. ydjN encodes an S-sulfocysteine transporter required by E. coli for growth on S-sulfocysteine as a sulfur source. FEMS Microbiol. Lett. 2016, 363, 17.
- Kredich, N.M.; Tomkins, G.M. The enzymic synthesis of l-cysteine in E. coli and Salmonella typhimurium. J. Biol. Chem. 1966, 241, 4955–4965.
- Ko, Y.-S.; Kim, J.W.; Lee, J.A.; Han, T.; Kim, J.B.; Park, J.E.; Lee, S.Y. Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem. Soc. Rev. 2020, 49, 4615–4636.
- Hoek, S.; Borodina, I. Transporter engineering in microbial cell factories: The ins, the outs, and the in-betweens. Curr. Opin. Biotechnol. 2020, 66, 186–194.
- Zhu, Y.; Zhou, C.; Wang, Y.; Li, C. Transporter engineering for microbial manufacturing. Biotechnol. J. 2020, 15, 1900494.
- Taniguchi, H.; Okano, K.; Honda, K. Modules for invitro metabolic engineering: Pathway assembly for bio-based production of value-added chemicals. Synth. Syst. Biotechnol. 2017, 2, 65–74.
- Wilding, K.M.; Schinn, S.-M.; Long, E.A.; Bundy, B.C. The emerging impact of cell-free chemical biosynthesis. Curr. Opin. Biotechnol. 2018, 53, 115–121.
- Hanatani, Y.; Imura, M.; Taniguchi, H.; Okano, K.; Toya, Y.; Iwakiri, R.; Honda, K. In vitro production of cysteine from glucose. Appl. Microbiol. Biotechnol. 2019, 103, 8009–8019.
- Imura, M.; Etoh, S.; Iwakiri, R.; Okano, K.; Honda, K. Improvement of production yield of L-cysteine through in vitro metabolic pathway with thermophilic enzymes. J. Biosic. Bioeng. 2021, 132, 585–591.
- Sprenger, G.A. Engineering of Microorganisms for the Production of Chemicals and Fuels from Renewable Resources; Springer Nat. Verl.: Berlin/Heidelberg, Germany, 2017; Volume 4, pp. 93–123.
- Luo, X.; Ge, X.; Cui, S.; Li, Y. Value-added processing of crude glycerol into chemicals and polymers. Bioresour. Technol. 2016, 215, 144–154.
- Gottlieb, K.; Albermann, C.; Sprenger, G.A. Improvement of L-phenylalanine production from glycerol by recombinant E. coli strains: The role of extra copies of glpK, glpX, and tktA genes. Microb. Cell Fact. 2014, 13, 96.
- Nguyen-Vo, T.P.; Liang, Y.; Sankaranarayanan, M.; Seol, E.; Chun, A.Y.; Ashok, S.; Chauhan, A.S.; Kim, J.R.; Park, S. Development of 3-hydroxypropionic-acid-tolerant strain of E. coli W and role of minor global regulator yieP. Metab. Eng. 2019, 53, 48–58.
- Zhang, X.; Zhang, D.; Zhu, J.F.; Liu, W.; Xu, G.Q.; Zhang, X.M.; Shi, J.S.; Xu, Z.H. High-yield production of L-serine from glycerol by engineered E. coli. J. Ind. Microbiol. Biotechnol. 2019, 46, 883–885.
- Rennig, M.; Mundhada, H.; Wordofa, G.G.; Gerngross, D.; Wulff, T.; Worberg, A.; Nielsen, A.T.; Nørholm, M.H.H. Industrializing a Bacterial Strain for l-Serine Production through Translation Initiation Optimization. ACS Synth. Biol. 2019, 8, 2347–2358.
- Duan, Y.; Zhai, W.; Liu, W.; Zhang, X.M.; Shi, J.S.; Xu, Z.H. Fine-Tuning Multi-Gene Clusters via Well-Characterized Gene Expression Regulatory Elements: Case Study of the Arginine Synthesis Pathway in C. glutamicum. ACS Synth. Biol. 2021, 10, 38–48.
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771.
- Cheng, F.; Tang, X.L.; Kardashliev, T. Transcription Factor-Based Biosensors in High-Throughput Screening: Advances and Applications. Biotechnol. J. 2018, 13, 1700648.
- Jiang, Y.; Qian, F.; Yang, J.; Liu, Y.M.; Dong, F.; Xu, C.M.; Sun, B.B.; Chen, B.; Xu, X.S.; Li, Y.; et al. CRISPR-Cpf1 assisted genome editing of C. glutamicum. Nat. Commun. 2017, 8, 15179.
- Zhang, J.; Yang, F.Y.; Yang, Y.P.; Jiang, Y.; Huo, Y.X. Optimizing a CRISPR-Cpf1-based genome engineering system for C. glutamicum. Microb. Cell Fact. 2019, 18, 60.
- Li, M.Y.; Chen, J.Z.; Wang, Y.; Liu, J.; Huang, J.W.; Chen, N.; Zheng, P.; Sun, J.B. Efficient Multiplex Gene Repression by CRISPR-dCpf1 in C. glutamicum. Front. Bioeng. Biotechnol. 2020, 8, 357.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.6K
Revisions:
2 times
(View History)
Update Date:
28 Sep 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





