
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Helen Onyeaka | -- | 3071 | 2023-09-14 10:30:23 | | | |
| 2 | Lindsay Dong | Meta information modification | 3071 | 2023-09-15 03:23:24 | | | | |
| 3 | Helen Onyeaka | Meta information modification | 3071 | 2024-06-26 13:31:03 | | |
Video Upload Options
Listeria monocytogenes (LM) is a bacterial pathogen responsible for listeriosis, a foodborne illness associated with high rates of mortality (20–30%) and hospitalisation. It is particularly dangerous among vulnerable groups, such as newborns, pregnant women and the elderly. The persistence of this organism in food-associated environments for months to years has been linked to several devastating listeriosis outbreaks. It may also result in significant costs to food businesses and economies.
1. Introduction
2. LM Biofilms
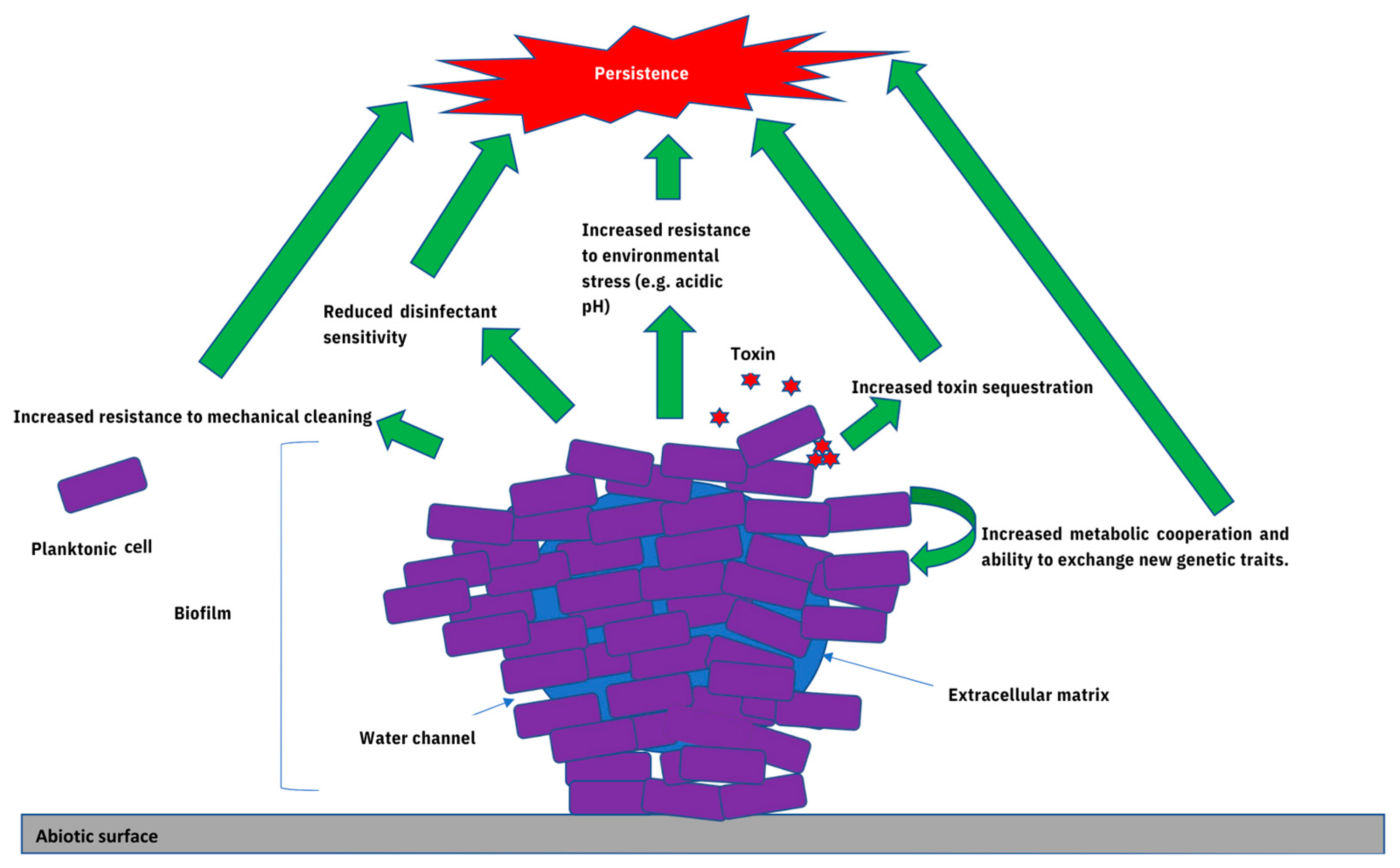
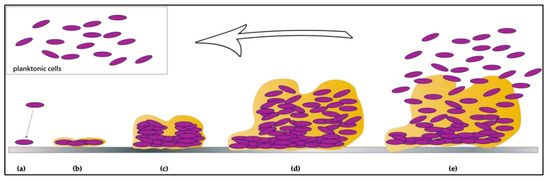
3. The Contribution of Biofilms towards Microbial Persistence within Food-Associated Environments
4. Phenotypic Comparisons of Biofilm Development in Persistent and PNP LM Strains
Several studies have reported evidence of enhanced biofilm formation among persistent LM strains [19][24][26][36][58][59][60][61][62]. For example, Norwood and Gilmour [24] measured the adherence to stainless steel coupons of 32 persistent and 25 PNP strains isolated from farms and food processing plants. After 24 h of surface contact at 25 °C, mean plate counts on the coupons were significantly higher for the persistent strains. Similar results were obtained by Lunden et al. [58] after they compared the adherence of a persistent strain to that of three PNP strains isolated from meat dicing plants on stainless steel coupons over two hours at 25 °C using epifluorescence microscopy counts.
A similar number of studies have reported no apparent difference between the biofilm formation of persistent and PNP LM strains [43][63][64][65][66][67][68][69][70][71][72][73]. For example, Djordjevic et al. [63] compared the biofilm formation of five persistent and five PNP strains isolated from fish processing plants to PVC microtiter plates using the crystal-violet method; after 40 h of surface contact at 32 °C, no relationship was observed between the biofilm levels of the persistent and PNP strains.
Factors that may explain the inconsistent findings between studies could include differences in experimental conditions, such as incubation temperature, surface material and nutrient availability [1][13][16][37].
The biofilm formation of persistent LM strains has also been found to vary according to the composition and concentration of the growth medium [19][40][74] and the surface material [75]. For example, Lee et al. [40] found that persistent strains exhibited significantly increased biofilm formation compared with PNP strains, but only in neat (undiluted), as opposed to dilute (1:10), brain heart infusion (BHI) broth.
In addition to different experimental conditions, there are several other factors that might account for the conflicting results between studies investigating the relationship between biofilm formation and persistence. These include differences in the choice of biofilm assay, sample size, persistence definitions, and/or test strain properties, such as lineage or serotype [4][26]. Differences in biofilm formation according to serotype have been reported in E-coli, with serotype O103:H2 isolates shown to produce more biofilm at 12 °C, 20 °C and 37 °C, compared with strains representative of serotypes O26:H11 or O103:H25 [76].
5. Genetic Mechanisms Involved in Biofilm Development in LM
6. WGS Comparisons of Biofilm Production in Persistent and PNP LM Strains
7. Conclusions
References
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes Biofilms in the Wonderland of Food Industry. Pathogens 2017, 6, 41.
- Mazaheri, T.; Cervantes-Huamán, B.R.H.; Bermúdez-Capdevila, M.; Ripolles-Avila, C.; Rodríguez-Jerez, J.J. Listeria monocytogenes Biofilms in the Food Industry: Is the Current Hygiene Program Sufficient to Combat the Persistence of the Pathogen? Microorganisms 2021, 9, 181.
- Chlebicz, A.; Śliżewska, K. Campylobacteriosis, Salmonellosis, Yersiniosis, and Listeriosis as Zoonotic Foodborne Diseases: A Review. Int. J. Environ. Res. Public Health 2018, 15, 863.
- Unrath, N.; McCabe, E.; Macori, G.; Fanning, S. Application of Whole Genome Sequencing to Aid in Deciphering the Persistence Potential of Listeria monocytogenes in Food Production Environments. Microorganisms 2021, 9, 1856.
- Lyytikäinen, O.; Autio, T.; Maijala, R.; Ruutu, P.; Honkanen-Buzalski, T.; Miettinen, M.; Hatakka, M.; Mikkola, J.; Anttila, V.-J.; Johansson, T.; et al. An Outbreak of Listeria Monocytogenes Serotype 3a Infections from Butter in Finland. J. Infect. Dis. 2000, 181, 1838–1841.
- Thomas, J.; Thomas, T.; Govender, N.; Govender, N.P.; McCarthy, K.M.; Erasmus, L.K.; Doyle, T.J.; Allam, M.; Ismail, A.; Ramalwa, N.; et al. Outbreak of Listeriosis in South Africa Associated with Processed Meat. N. Engl. J. Med. 2020, 382, 632–643.
- McLauchlin, J.; Grant, K.A.; Amar, C.F.L. Human foodborne listeriosis in England and Wales, 1981 to 2015. Epidemiol. Infect. 2020, 148, e54.
- Malley, T.J.V.; Butts, J.; Wiedmann, M. Seek and destroy process: Listeria monocytogenes process controls in the ready-to-eat meat and poultry industry. J. Food Prot. 2015, 78, 436–445.
- EFSA/ECDC. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e06971.
- de Noordhout, C.M.M.; Devleesschauwer, B.M.; Angulo, F.J.P.; Verbeke, G.P.; Haagsma, J.P.; Kirk, M.P.; Havelaar, A.P.; Speybroeck, N.P. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 1073–1082.
- Osek, J.; Lachtara, B.; Wieczorek, K. How This Pathogen Survives in Food-Production Environments? Front. Microbiol. 2022, 13, 866462.
- Kathariou, S. Listeria monocytogenes Virulence and Pathogenicity, a Food Safety Perspective. J. Food Prot. 2002, 65, 1811–1829.
- Valderrama, W.B.; Cutter, C.N. An Ecological Perspective of Listeria monocytogenes Biofilms in Food Processing Facilities. Crit. Rev. Food Sci. Nutr. 2013, 53, 801–817.
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185.
- Tompkin, R.B. Control of Listeria monocytogenes in the Food-Processing Environment. J. Food Prot. 2002, 65, 709–725.
- Møretrø, T.; Langsrud, S. Listeria monocytogenes: Biofilm formation and persistence in food-processing environments. Biofilms 2004, 1, 107–121.
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8.
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes Persistence in Food-Associated Environments: Epidemiology, Strain Characteristics, and Implications for Public Health. J. Food Prot. 2014, 77, 150–170.
- Natalia, W.-K.; Ewa, W.-Z.; Krzysztof, S.; Agnieszka, K.; Zuzanna, B.; Justyna, B.-K.; Zuzanna, K.; Eugenia, G.-K. Comparison of Selected Phenotypic Features of Persistent and Sporadic Strains of Listeria monocytogenes Sampled from Fish Processing Plants. Foods 2022, 11, 1492.
- Orsi, R.H.; Borowsky, M.L.; Lauer, P.; Young, S.K.; Nusbaum, C.; Galagan, J.E.; Birren, B.W.; Ivy, R.A.; Sun, Q.; Graves, L.M.; et al. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genom. 2008, 9, 539.
- Forauer, E.; Wu, S.T.; Etter, A.J. Listeria monocytogenes in the retail deli environment: A review. Food Control 2021, 119, 107443.
- Holah, J.T.; Taylor, J.H.; Dawson, D.J.; Hall, K.E. Biocide use in the food industry and the disinfectant resistance of persistent strains of Listeria monocytogenes and Escherichia coli. Symp. Ser. (Soc. Appl. Microbiol.) 2002, 92, 111S–120S.
- Wulff, G.; Gram, L.; Ahrens, P.; Vogel, B.F. One Group of Genetically Similar Listeria monocytogenes Strains Frequently Dominates and Persists in Several Fish Slaughter- and Smokehouses. Appl. Environ. Microbiol. 2006, 72, 4313–4322.
- Norwood, D.E.; Gilmour, A. Adherence of Listeria monocytogenes strains to stainless steel coupons. J. Appl. Microbiol. 1999, 86, 576–582.
- Lunden, J.M.; Miettinen, M.K.; Autio, T.J.; Korkeala, H.J. Persistent Listeria monocytogenes Strains Show Enhanced Adherence to Food Contact Surface after Short Contact Times. J. Food Prot. 2000, 63, 1204–1207.
- Borucki, M.K.; Peppin, J.D.; White, D.; Loge, F.; Call, D.R. Variation in Biofilm Formation among Strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7336–7342.
- Monds, R.D.; O’Toole, G.A. The developmental model of microbial biofilms: Ten years of a paradigm up for review. Trends Microbiol. 2008, 17, 73–87.
- Marsh, E.J.; Luo, H.; Wang, H. A three-tiered approach to differentiate Listeria monocytogenes biofilm-forming abilities. FEMS Microbiol. Lett. 2003, 228, 203–210.
- Rieu, A.; Briandet, R.; Habimana, O.; Garmyn, D.; Guzzo, J.; Piveteau, P. Listeria monocytogenes EGD-e Biofilms: No Mushrooms but a Network of Knitted Chains. Appl. Environ. Microbiol. 2008, 74, 4491–4497.
- Milanov, D.; Ašanin, R.; Vidić, R.; Katić, V.; Plavša, N. Examination of the capabilities of attachment and biofilm formation of different Listeria monocytogenes STRAINS. Biotechnol. Anim. Husb. 2009, 5–6, 1255–1265.
- Blackman, I.C.; Frank, J.F. Growth of Listeria monocytogenes as a biofilm on various food-processing surfaces. J. Food Prot. 1996, 59, 827–831.
- Chavant, P.; Martinie, B.; Thierry, M.; Marie-Noëlle, B.-F.; Michel, H. Listeria monocytogenes LO28: Surface Physicochemical Properties and Ability To Form Biofilms at Different Temperatures and Growth Phases. Appl. Environ. Microbiol. 2002, 68, 728–737.
- Colagiorgi, A.; Di Ciccio, P.; Zanardi, E.; Ghidini, S.; Ianieri, A. A Look inside the Listeria monocytogenes Biofilms Extracellular Matrix. Microorganisms 2016, 4, 22.
- Chmielewski, R.A.N.; Frank, J.F. Biofilm Formation and Control in Food Processing Facilities. Compr. Rev. Food Sci. Food Saf. 2003, 2, 22–32.
- Kumar, C.G.; Anand, S.K. Significance of microbial biofilms in food industry: A review. Int. J. Food Microbiol. 1998, 42, 9–27.
- Latorre, A.A.; Van Kessel, J.A.S.; Karns, J.S.; Zurakowski, M.J.; Pradhan, A.K.; Boor, K.J.; Adolph, E.; Sukhnanand, S.; Schukken, Y.H. Increased In Vitro Adherence and On-Farm Persistence of Predominant and Persistent Listeria monocytogenes Strains in the Milking System. Appl. Environ. Microbiol. 2011, 77, 3676–3684.
- Russo, P.; Hadjilouka, A.; Beneduce, L.; Capozzi, V.; Paramithiotis, S.; Drosinos, E.H.; Spano, G. Effect of different conditions on Listeria monocytogenes biofilm formation and removal. Czech J. Food Sci. 2018, 36, 208–214.
- Duffy, G.; Sheridan, J.J. The effect of temperature, pH and medium in a surface adhesion immunofluorescent technique for detection of Listeria monocytogenes. J. Appl. Microbiol. 1997, 83, 95–101.
- Briandet, R.; Leriche, V.; Carpentier, B.; Bellon-Fontaine, M.-N. Effects of the Growth Procedure on the Surface Hydrophobicity of Listeria monocytogenes Cells and Their Adhesion to Stainless Steel. J. Food Prot. 1999, 62, 994–998.
- Lee, B.-H.; Cole, S.; Badel-Berchoux, S.; Guillier, L.; Felix, B.; Krezdorn, N.; Hébraud, M.; Bernardi, T.; Sultan, I.; Piveteau, P. Biofilm Formation of Listeria monocytogenes Strains Under Food Processing Environments and Pan-Genome-Wide Association Study. Front. Microbiol. 2019, 10, 2698.
- Di Bonaventura, G.; Piccolomini, R.; Paludi, D.; D’Orio, V.; Vergara, A.; Conter, M.; Ianieri, A. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 2008, 104, 1552–1561.
- Kadam, S.R.; den Besten, H.M.W.; van der Veen, S.; Zwietering, M.H.; Moezelaar, R.; Abee, T. Diversity assessment of Listeria monocytogenes biofilm formation: Impact of growth condition, serotype and strain origin. Int. J. Food Microbiol. 2013, 165, 259–264.
- Nilsson, R.E.; Ross, T.; Bowman, J.P. Variability in biofilm production by Listeria monocytogenes correlated to strain origin and growth conditions. Int. J. Food Microbiol. 2011, 150, 14–24.
- Smoot, L.M.; Pierson, M.D. Influence of Environmental Stress on the Kinetics and Strength of Attachment of Listeria monocytogenes Scott A to Buna-N Rubber and Stainless Steel. J. Food Prot. 1998, 61, 1286–1292.
- Bereksi, N.; Gavini, F.; Bénézech, T.; Faille, C. Growth, morphology and surface properties of Listeria monocytogenes Scott A and LO28 under saline and acid environments. J. Appl. Microbiol. 2002, 92, 556–565.
- Mafu, A.A.; Roy, D.; Goulet, J.; Savoie, L. Characterization of physicochemical forces involved in adhesion of Listeria monocytogenes to surfaces. Appl. Environ. Microbiol. 1991, 57, 1969–1973.
- Coughlan, L.M.; Cotter, P.D.; Hill, C.; Alvarez-Ordóñez, A. New weapons to fight old enemies: Novel strategies for the (bio)control of bacterial biofilms in the food industry. Front. Microbiol. 2016, 7, 1641.
- Ripolles-Avila, C.; Hascoët, A.S.; Guerrero-Navarro, A.E.; Rodríguez-Jerez, J.J. Establishment of incubation conditions to optimize the in vitro formation of mature Listeria monocytogenes biofilms on food-contact surfaces. Food Control 2018, 92, 240–248.
- Davey, M.E.; O’Toole, G.A. Microbial Biofilms: From Ecology to Molecular Genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867.
- Bai, X.; Nakatsu, C.H.; Bhunia, A.K. Bacterial Biofilms and Their Implications in Pathogenesis and Food Safety. Foods 2021, 10, 2117.
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; De Keersmaecker, S.C.J. Salmonella biofilms: An overview on occurrence, structure, regulation and eradication. Food Res. Int. 2012, 45, 502–531.
- Vestby, L.K.; Møretrø, T.; Langsrud, S.; Heir, E.; Nesse, L.L. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal- and feed factories. BMC Vet. Res. 2009, 5, 20.
- Yang, X.; Tran, F.; Youssef, M.K.; Gill, C.O. Determination of Sources of Escherichia coli on Beef by Multiple-Locus Variable-Number Tandem Repeat Analysis. J. Food Prot. 2015, 78, 1296–1302.
- Wang, R.; Luedtke, B.E.; Bosilevac, J.M.; Schmidt, J.W.; Kalchayanand, N.; Arthur, T.M. Escherichia coli O157:H7 Strains Isolated from High-Event Period Beef Contamination Have Strong Biofilm-Forming Ability and Low Sanitizer Susceptibility, Which Are Associated with High pO157 Plasmid Copy Number. J. Food Prot. 2016, 79, 1875–1883.
- Alvarez-Ordóñez, A.; Coughlan, L.M.; Briandet, R.; Cotter, P.D. Biofilms in Food Processing Environments: Challenges and Opportunities. Annu. Rev. Food Sci. Technol. 2019, 10, 173–195.
- Lamas, A.; Regal, P.; Vázquez, B.; Miranda, J.M.; Cepeda, A.; Franco, C.M. Salmonella and Campylobacter biofilm formation: A comparative assessment from farm to fork. J. Sci. Food Agric. 2018, 98, 4014–4032.
- Larsen, M.H.; Dalmasso, M.; Ingmer, H.; Langsrud, S.; Malakauskas, M.; Mader, A.; Møretrø, T.; Smole Možina, S.; Rychli, K.; Wagner, M.; et al. Persistence of foodborne pathogens and their control in primary and secondary food production chains. Food Control 2014, 44, 92–109.
- Lunden, J.M.; Autio, T.J.; Korkeala, H.J. Transfer of Persistent Listeria monocytogenes Contamination between Food-Processing Plants Associated with a Dicing Machine. J. Food Prot. 2002, 65, 1129–1133.
- Poimenidou, S.V.; Chrysadakou, M.; Tzakoniati, A.; Bikouli, V.C.; Nychas, G.-J.; Skandamis, P.N. Variability of Listeria monocytogenes strains in biofilm formation on stainless steel and polystyrene materials and resistance to peracetic acid and quaternary ammonium compounds. Int. J. Food Microbiol. 2016, 237, 164–171.
- Nowak, J.; Cruz, C.D.; Tempelaars, M.; Abee, T.; van Vliet, A.H.M.; Fletcher, G.C.; Hedderley, D.; Palmer, J.; Flint, S. Persistent Listeria monocytogenes strains isolated from mussel production facilities form more biofilm but are not linked to specific genetic markers. Int. J. Food Microbiol. 2017, 256, 45–53.
- Rodríguez-Campos, D.; Rodríguez-Melcón, C.; Alonso-Calleja, C.; Capita, R. Persistent Listeria monocytogenes isolates from a poultry-processing facility form more biofilm but do not have a greater resistance to disinfectants than sporadic strains. Pathogens 2019, 8, 250.
- Harrand, A.S.; Jagadeesan, B.; Baert, L.; Wiedmann, M.; Orsi, R.H. Evolution of Listeria monocytogenes in a Food Processing Plant Involves Limited Single-Nucleotide Substitutions but Considerable Diversification by Gain and Loss of Prophages. Appl. Environ. Microbiol. 2020, 86, e02493-19.
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L.A. Microtiter Plate Assay for Assessment of Listeria monocytogenes Biofilm Formation. Appl. Environ. Microbiol. 2002, 68, 2950–2958.
- Harvey, J.; Keenan, K.P.; Gilmour, A. Assessing biofilm formation by Listeria monocytogenes strains. Food Microbiol. 2007, 24, 380–392.
- Jensen, A.; Larsen, M.H.; Ingmer, H.; Vogel, B.F.; Gram, L. Sodium Chloride Enhances Adherence and Aggregation and Strain Variation Influences Invasiveness of Listeria monocytogenes Strains. J. Food Prot. 2007, 70, 592–599.
- Cruz, C.D.; Fletcher, G.C. Prevalence and biofilm-forming ability of Listeria monocytogenes in New Zealand mussel (Perna canaliculus) processing plants. Food Microbiol. 2011, 28, 1387–1393.
- Szlavik, J.; Paiva, D.S.; Mørk, N.; van den Berg, F.; Verran, J.; Whitehead, K.; Knøchel, S.; Nielsen, D.S. Initial adhesion of Listeria monocytogenes to solid surfaces under liquid flow. Int. J. Food Microbiol. 2012, 152, 181–188.
- Costa, A.; Bertolotti, L.; Brito, L.; Civera, T. Biofilm Formation and Disinfectant Susceptibility of Persistent and Nonpersistent Listeria monocytogenes Isolates from Gorgonzola Cheese Processing Plants. Foodborne Pathog. Dis. 2016, 13, 602–609.
- Korenova, J.; Oravcova, K.; Veghova, J.; Karpiskova, R.; Kuchta, T. Biofilm formation in various conditions is not a key factor of persistence potential of Listeria monocytogenes in food-processing environment. J. Food. Nutr. Res 2016, 55, 189–193.
- In Lee, S.H.; Barancelli, G.V.; de Camargo, T.M.; Corassin, C.H.; Rosim, R.E.; da Cruz, A.G.; Cappato, L.P.; de Oliveira, C.A.F. Biofilm-producing ability of Listeria monocytogenes isolates from Brazilian cheese processing plants. Food Res. Int. 2017, 91, 88–91.
- Cherifi, T.; Carrillo, C.; Lambert, D.; Miniaï, I.; Quessy, S.; Larivière-Gauthier, G.; Blais, B.; Fravalo, P. Genomic characterization of Listeria monocytogenes isolates reveals that their persistence in a pig slaughterhouse is linked to the presence of benzalkonium chloride resistance genes. BMC Microbiol. 2018, 18, 220.
- Sereno, M.J.; Viana, C.; Pegoraro, K.; da Silva, D.A.L.; Yamatogi, R.S.; Nero, L.A.; Bersot, L.d.S. Distribution, adhesion, virulence and antibiotic resistance of persistent Listeria monocytogenes in a pig slaughterhouse in Brazil. Food Microbiol. 2019, 84, 103234.
- Ramires, T.; Kleinubing, N.R.; Iglesias, M.A.; Vitola, H.R.S.; Núncio, A.S.P.; Kroning, I.S.; Moreira, G.M.S.G.; Fiorentini, Â.M.; da Silva, W.P. Genetic diversity, biofilm and virulence characteristics of Listeria monocytogenes in salmon sushi. Food Res. Int. 2021, 140, 109871.
- da Silva, D.A.L.; de Melo Tavares, R.; Camargo, A.C.; Yamatogi, R.S.; De Martinis, E.C.P.; Nero, L.A. Biofilm growth by Listeria monocytogenes on stainless steel and expression of biofilm-related genes under stressing conditions. World J. Microbiol. Biotechnol. 2021, 37, 119.
- Magalhaes, R.; Ferreira, V.; Biscottini, G.; Brandao, T.R.S.; Almeida, G.; Teixeira, P. Biofilm formation by persistent and non-persistent listeria monocytogenes strains on abiotic surfaces. Acta Aliment. 2017, 46, 43–50.
- Nesse, L.L.; Sekse, C.; Berg, K.; Johannesen, K.C.; Solheim, H.; Vestby, L.K.; Urdahl, A.M. Potentially pathogenic Escherichia coli can form a biofilm under conditions relevant to the food production chain. Appl. Environ. Microbiol. 2014, 80, 2042–2049.
- Renier, S.; Hébraud, M.; Desvaux, M. Molecular biology of surface colonization by Listeria monocytogenes: An additional facet of an opportunistic Gram-positive foodborne pathogen. Environ. Microbiol. 2011, 13, 835–850.
- Vatanyoopaisarn, S.; Aisha, N.; Christine, E.R.D.; Catherine, E.D.R.; Will, M.W. Effect of Flagella on Initial Attachment of Listeria monocytogenes to Stainless Steel. Appl. Environ. Microbiol. 2000, 66, 860–863.
- Lemon, K.P.; Higgins, D.E.; Kolter, R. Flagellar Motility Is Critical for Listeria monocytogenes Biofilm Formation. J. Bacteriol. 2007, 189, 4418–4424.
- Kumar, S.; Parvathi, A.; George, J.; Krohne, G.; Karunasagar, I.; Karunasagar, I. A study on the effects of some laboratory-derived genetic mutations on biofilm formation by Listeria monocytogenes. World J. Microbiol. Biotechnol. 2008, 25, 527–531.
- Todhanakasem, T.; Young, G.M. Loss of Flagellum-Based Motility by Listeria monocytogenes Results in Formation of Hyperbiofilms. J. Bacteriol. 2008, 190, 6030–6034.
- Lemon, K.P.; Freitag, N.E.; Kolter, R. The Virulence Regulator PrfA Promotes Biofilm Formation by Listeria monocytogenes. J. Bacteriol. 2010, 192, 3969–3976.
- Zhou, Q.; Feng, F.; Wang, L.; Feng, X.; Yin, X.; Luo, Q. Virulence Regulator PrfA is Essential for Biofilm Formation in Listeria monocytogenes but not in Listeria innocua. Curr. Microbiol. 2011, 63, 186–192.
- Knudsen, G.M.; Olsen, J.E.; Dons, L. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol. Lett. 2004, 240, 171–179.
- Gueriri, I.; Cyncynatus, C.; Dubrac, S.; Arana, A.T.; Dussurget, O.; Msadek, T. The DegU orphan response regulator of Listeria monocytogenes autorepresses its own synthesis and is required for bacterial motility, virulence and biofilm formation. Microbiology 2008, 154, 2251–2264.
- Travier, L.; Guadagnini, S.; Gouin, E.; Dufour, A.; Chenal-Francisque, V.; Cossart, P.; Olivo-Marin, J.-C.; Ghigo, J.-M.; Disson, O.; Lecuit, M. ActA promotes Listeria monocytogenes aggregation, intestinal colonization and carriage. PLoS Pathog. 2013, 9, e1003131.
- Schwab, U.; Hu, Y.; Wiedmann, M.; Boor, K.J. Alternative sigma factor sigma(B) is not essential for Listeria monocytogenes surface attachment. J. Food Prot. 2005, 68, 311–317.
- van der Veen, S.; Abee, T. Importance of SigB for Listeria monocytogenes Static and Continuous-Flow Biofilm Formation and Disinfectant Resistance. Appl. Environ. Microbiol. 2010, 76, 7854–7860.
- Janež, N.; Škrlj, B.; Sterniša, M.; Klančnik, A.; Sabotič, J. The role of the Listeria monocytogenes surfactome in biofilm formation. Microb. Biotechnol. 2021, 14, 1269–1281.
- Chen, B.Y.; Kim, T.J.; Jung, Y.S.; Silva, J.L. Attachment Strength of Listeria monocytogenes and its Internalin-Negative Mutants. Food Biophys. 2008, 3, 329–332.
- Chen, B.-Y.; Kim, T.-J.; Silva, J.L.; Jung, Y.-S. Positive Correlation Between the Expression of inlA and inlB Genes of Listeria monocytogenes and Its Attachment Strength on Glass Surface. Food Biophys. 2009, 4, 304–311.
- Franciosa, G.; Maugliani, A.; Scalfaro, C.; Floridi, F.; Aureli, P. Expression of Internalin A and Biofilm Formation among Listeria monocytogenes Clinical Isolates. Int. J. Immunopathol. Pharmacol. 2009, 22, 183–193.
- Popowska, M.; Krawczyk-Balska, A.; Ostrowski, R.; Desvaux, M. InlL from Listeria monocytogenes is involved in biofilm formation and adhesion to mucin. Front Microbiol. 2017, 8, 660.
- Rieu, A.; Weidmann, S.; Garmyn, D.; Piveteau, P.; Guzzo, J. agr System of Listeria monocytogenes EGD-e: Role in Adherence and Differential Expression Pattern. Appl. Environ. Microbiol. 2007, 73, 6125–6133.
- Riedel, C.U.; Monk, I.R.; Casey, P.G.; Waidmann, M.S.; Gahan, C.G.M.; Hill, C. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol. Microbiol. 2009, 71, 1177–1189.
- Zetzmann, M.; Bucur, F.I.; Crauwels, P.; Borda, D.; Nicolau, A.I.; Grigore-Gurgu, L.; Seibold, G.M.; Riedel, C.U. Characterization of the biofilm phenotype of a Listeria monocytogenes mutant deficient in agr peptide sensing. Microbiologyopen 2019, 8, e00826-n/a.
- Bonsaglia, E.C.R.; Silva, N.C.C.; Fernades Júnior, A.; Araújo Júnior, J.P.; Tsunemi, M.H.; Rall, V.L.M. Production of biofilm by Listeria monocytogenes in different materials and temperatures. Food Control 2014, 35, 386–391.
- Belval, S.C.; Gal, L.; Margiewes, S.; Garmyn, D.; Piveteau, P.; Guzzo, J. Assessment of the Roles of LuxS, S-Ribosyl Homocysteine, and Autoinducer 2 in Cell Attachment during Biofilm Formation by Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 2006, 72, 2644–2650.
- Sela, S.; Frank, S.; Belausov, E.; Pinto, R. A Mutation in the luxS Gene Influences Listeria monocytogenes Biofilm Formation. Appl. Environ. Microbiol. 2006, 72, 5653–5658.
- Nielsen, E.M.; Björkman, J.T.; Kiil, K.; Grant, K.; Dallman, T.; Painset, A.; Amar, C.; Roussel, S.; Guillier, L.; Félix, B.; et al. Closing gaps for performing a risk assessment on Listeria monocytogenes in ready-to-eat (RTE) foods: Activity 3, the comparison of isolates from different compartments along the food chain, and from humans using whole genome sequencing (WGS) analysis. EFSA Support. Publ. 2017, 14, 1151E.
- Muhterem-Uyar, M.; Ciolacu, L.; Wagner, K.-H.; Wagner, M.; Schmitz-Esser, S.; Stessl, B. New Aspects on Listeria monocytogenes ST5-ECVI Predominance in a Heavily Contaminated Cheese Processing Environment. Front. Microbiol. 2018, 9, 64.




