
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Satoru Matsuda | -- | 2045 | 2023-09-07 15:56:03 | | | |
| 2 | Peter Tang | Meta information modification | 2045 | 2023-09-08 04:47:52 | | |
Video Upload Options
Non-coding RNAs (ncRNAs) are indispensable for adjusting gene expression and genetic programming throughout development and for health as well as cardiovascular diseases. Cardiac arrhythmia is a frequent cardiovascular disease that has a complex pathology. Studies have shown that ncRNAs are also associated with cardiac arrhythmias. Many non-coding RNAs and/or genomes have been reported as genetic background for cardiac arrhythmias. In general, arrhythmias may be affected by several functional and structural changes in the myocardium of the heart. Therefore, ncRNAs might be indispensable regulators of gene expression in cardiomyocytes, which could play a dynamic role in regulating the stability of cardiac conduction and/or in the remodeling process.
1. Introduction
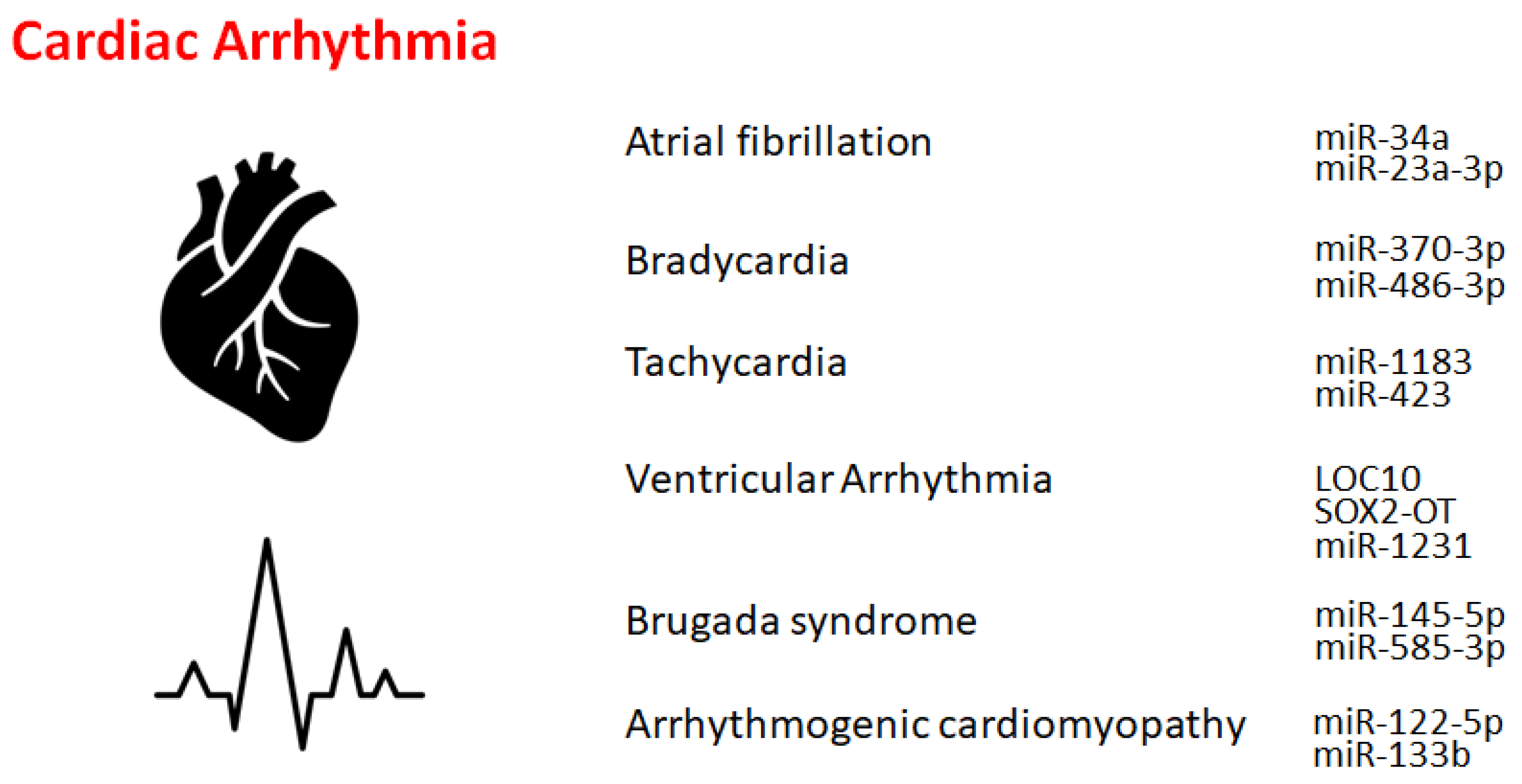
2. Atrial Fibrillation
3. Bradycardia and Tachycardia
4. Other Cardiac Rhythm Disorders
5. Gut Microbiota and Cardiac Arrhythmia
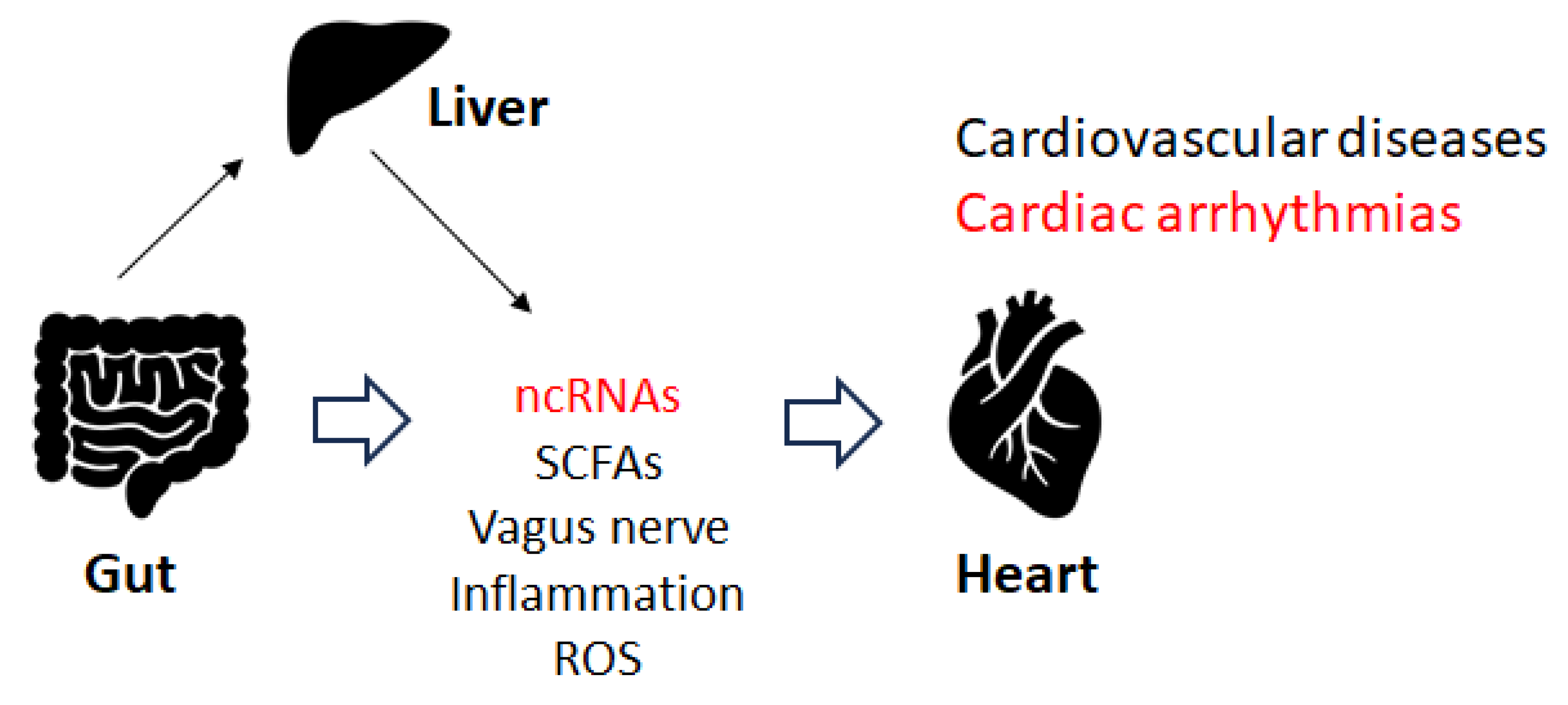
6. Immune Pathway and Cardiac Arrhythmias
References
- Fan, W.; Sun, X.; Yang, C.; Wan, J.; Luo, H.; Liao, B. Pacemaker activity and ion channels in the sinoatrial node cells: MicroRNAs and arrhythmia. Prog. Biophys. Mol. Biol. 2023, 177, 151.
- Yaniv, Y.; Lakatta, E.G.; Maltsev, V.A. From two competing oscillators to one coupled-clock pacemaker cell system. Front. Physiol. 2015, 6, 28.
- Wilde, A.A.; Bezzina, C.R. Genetics of cardiac arrhythmias. Heart 2005, 91, 1352.
- Kawaguchi, S.; Moukette, B.; Hayasaka, T.; Haskell, A.K.; Mah, J.; Sepúlveda, M.N.; Tang, Y.; Kim, I.M. Noncoding RNAs as Key Regulators for Cardiac Development and Cardiovascular Diseases. J. Cardiovasc. Dev. Dis. 2023, 10, 166.
- Sallam, T.; Sandhu, J.; Tontonoz, P. Long Noncoding RNA Discovery in Cardiovascular Disease: Decoding Form to Function. Circ. Res. 2018, 122, 155.
- Zhang, C.; Han, B.; Xu, T.; Li, D. The biological function and potential mechanism of long non-coding RNAs in cardiovascular disease. J. Cell. Mol. Med. 2020, 24, 12900.
- Correia, C.C.M.; Rodrigues, L.F.; de Avila Pelozin, B.R.; Oliveira, E.M.; Fernandes, T. Long Non-Coding RNAs in Cardiovascular Diseases: Potential Function as Biomarkers and Therapeutic Targets of Exercise Training. Non-Coding RNA 2021, 7, 65.
- Gudbjartsson, D.F.; Arnar, D.O.; Helgadottir, A.; Gretarsdottir, S.; Holm, H.; Sigurdsson, A.; Jonasdottir, A.; Baker, A.; Thorleifsson, G.; Kristjansson, K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 2007, 448, 353.
- Ruan, Z.B.; Wang, F.; Gongben, B.D.; Chen, G.C.; Zhu, L. Identification of Circulating lncRNA Expression Profiles in Patients with Atrial Fibrillation. Dis. Mark. 2020, 2020, 8872142.
- Xie, L.; Huang, G.L.; Gao, M.L.; Huang, J.L.; Li, H.L.; Xia, H.L.; Xiang, X.L.; Wu, S.L.; Ruan, Y. Identification of Atrial Fibrillation-Related lncRNA Based on Bioinformatic Analysis. Dis. Mark. 2022, 2022, 8307975.
- Barstow, C.; McDivitt, J.D. Cardiovascular Disease Update: Bradyarrhythmias. FP Essent. 2017, 454, 18.
- Yanni, J.; D’Souza, A.; Wang, Y.; Li, N.; Hansen, B.J.; Zakharkin, S.O.; Smith, M.; Hayward, C.; Whitson, B.A.; Mohler, P.J.; et al. Silencing miR-370-3p rescues funny current and sinus node function in heart failure. Sci. Rep. 2020, 10, 11279.
- Petkova, M.; Atkinson, A.J.; Yanni, J.; Stuart, L.; Aminu, A.J.; Ivanova, A.D.; Pustovit, K.B.; Geragthy, C.; Feather, A.; Li, N.; et al. Identification of Key Small Non-Coding MicroRNAs Controlling Pacemaker Mechanisms in the Human Sinus Node. J. Am. Heart Assoc. 2020, 9, e016590.
- Aminu, A.J.; Petkova, M.; Atkinson, A.J.; Yanni, J.; Morris, A.D.; Simms, R.T.; Chen, W.; Yin, Z.; Kuniewicz, M.; Holda, M.K.; et al. Further insights into the molecular complexity of the human sinus node—The role of ‘novel’ transcription factors and microRNAs. Prog. Biophys. Mol. Biol. 2021, 166, 86.
- Zhao, H.; Tan, Z.; Zhou, J.; Wu, Y.; Hu, Q.; Ling, Q.; Ling, J.; Liu, M.; Ma, J.; Zhang, D.; et al. The regulation of circRNA and lncRNAprotein binding in cardiovascular diseases: Emerging therapeutic targets. Biomed. Pharmacother. 2023, 165, 115067.
- Liang, Y.; Wang, B.; Huang, H.; Wang, M.; Wu, Q.; Zhao, Y.; He, Y. Silenced SOX2-OT alleviates ventricular arrhythmia associated with heart failure by inhibiting NLRP3 expression via regulating miR-2355-3p. Immun. Inflamm. Dis. 2021, 9, 255.
- Xiong, F.; Mao, R.; Zhang, L.; Zhao, R.; Tan, K.; Liu, C.; Xu, J.; Du, G.; Zhang, T. CircNPHP4 in monocyte-derived small extracellular vesicles controls heterogeneous adhesion in coronary heart atherosclerotic disease. Cell Death Dis. 2021, 12, 948.
- Shi, Y.; Qiao, L.; Han, F.; Xie, X.; Wang, W. MiR-1231 regulates L-calcium in ventricular arrhythmia in chronic heart failure. Minerva Med. 2021, 112, 305.
- Sarquella-Brugada, G.; Cesar, S.; Zambrano, M.D.; Fernandez-Falgueras, A.; Fiol, V.; Iglesias, A.; Torres, F.; Garcia-Algar, O.; Arbelo, E.; Brugada, J.; et al. Electrocardiographic Assessment and Genetic Analysis in Neonates: A Current Topic of Discussion. Curr. Cardiol. Rev. 2019, 15, 30.
- Lucking, E.F.; O’Connor, K.M.; Strain, C.R.; Fouhy, F.; Bastiaanssen, T.F.S.; Burns, D.P.; Golubeva, A.V.; Stanton, C.; Clarke, G.; Cryan, J.F.; et al. Chronic intermittent hypoxia disrupts cardiorespiratory homeostasis and gut microbiota composition in adult male guinea-pigs. eBioMedicine 2018, 38, 191.
- Anthony, W.E.; Wang, B.; Sukhum, K.V.; D’Souza, A.W.; Hink, T.; Cass, C.; Seiler, S.; Reske, K.A.; Coon, C.; Dubberke, E.R.; et al. Acute and persistent effects of commonly used antibiotics on the gut microbiome and resistome in healthy adults. Cell Rep. 2022, 39, 110649.
- Tierney, B.T.; Yang, Z.; Luber, J.M.; Beaudin, M.; Wibowo, M.C.; Baek, C.; Mehlenbacher, E.; Patel, C.J.; Kostic, A.D. The Landscape of Genetic Content in the Gut and Oral Human Microbiome. Cell Host Microbe 2019, 26, 283.
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212.
- Lal, S.; Kirkup, A.J.; Brunsden, A.M.; Thompson, D.G.; Grundy, D. Vagal afferent responses to fatty acids of different chain length in the rat. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 281, G907.
- Onyszkiewicz, M.; Gawrys-Kopczynska, M.; Konopelski, P.; Aleksandrowicz, M.; Sawicka, A.; Koźniewska, E.; Samborowska, E.; Ufnal, M. Butyric acid, a gut bacteria metabolite, lowers arterial blood pressure via colon-vagus nerve signaling and GPR41/43 receptors. Pflug. Arch. 2019, 471, 1441.
- Tabata, T.; Yamashita, T.; Hosomi, K.; Park, J.; Hayashi, T.; Yoshida, N.; Saito, Y.; Fukuzawa, K.; Konishi, K.; Murakami, H.; et al. Gut microbial composition in patients with atrial fibrillation: Effects of diet and drugs. Heart Vessels 2021, 36, 105.
- Celikyurt, I.; Meier, C.R.; Kühne, M.; Schaer, B. Safety and Interactions of Direct Oral Anticoagulants with Antiarrhythmic Drugs. Drug Saf. 2017, 40, 1091.
- Aliabadi, T.; Saberi, E.A.; Motameni Tabatabaei, A.; Tahmasebi, E. Antibiotic use in endodontic treatment during pregnancy: A narrative review. Eur. J. Transl. Myol. 2022, 32, 10813.
- Valdivielso, J.M.; Balafa, O.; Ekart, R.; Ferro, C.J.; Mallamaci, F.; Mark, P.B.; Rossignol, P.; Sarafidis, P.; Del Vecchio, L.; Ortiz, A. Hyperkalemia in Chronic Kidney Disease in the New Era of Kidney Protection Therapies. Drugs 2021, 81, 1467.
- Fardi, F.; Khasraghi, L.B.; Shahbakhti, N.; Salami Naseriyan, A.; Najafi, S.; Sanaaee, S.; Alipourfard, I.; Zamany, M.; Karamipour, S.; Jahani, M.; et al. An interplay between non-coding RNAs and gut microbiota in human health. Diabetes Res. Clin. Pract. 2023, 201, 110739.
- Zhou, F.; Wang, X.; Wang, L.; Sun, X.; Tan, G.; Wei, W.; Zheng, G.; Ma, X.; Tian, D.; Yu, H. Genetics, Epigenetics, Cellular Immunology, and Gut Microbiota: Emerging Links with Graves’ Disease. Front. Cell Dev. Biol. 2022, 9, 794912.
- Wang, X.; Fan, H.; Wang, Y.; Yin, X.; Liu, G.; Gao, C.; Li, X.; Liang, B. Elevated Peripheral T Helper Cells Are Associated with Atrial Fibrillation in Patients with Rheumatoid Arthritis. Front. Immunol. 2021, 12, 744254.
- He, Y.; Chen, X.; Guo, X.; Yin, H.; Ma, N.; Tang, M.; Liu, H.; Mei, J. Th17/Treg Ratio in Serum Predicts Onset of Postoperative Atrial Fibrillation After Off-Pump Coronary Artery Bypass Graft Surgery. Heart Lung Circ. 2018, 27, 1467.
- Jia, N.; Lin, X.; Ma, S.; Ge, S.; Mu, S.; Yang, C.; Shi, S.; Gao, L.; Xu, J.; Bo, T.; et al. Amelioration of hepatic steatosis is associated with modulation of gut microbiota and suppression of hepatic miR-34a in Gynostemma pentaphylla (Thunb.) Makino treated mice. Nutr. Metab. 2018, 15, 86.
- Li, L.; Li, C.; Lv, M.; Hu, Q.; Guo, L.; Xiong, D. Correlation between alterations of gut microbiota and miR-122-5p expression in patients with type 2 diabetes mellitus. Ann. Transl. Med. 2020, 8, 1481.
- Bueno Marinas, M.; Celeghin, R.; Cason, M.; Bariani, R.; Frigo, A.C.; Jager, J.; Syrris, P.; Elliott, P.M.; Bauce, B.; Thiene, G.; et al. A microRNA Expression Profile as Non-Invasive Biomarker in a Large Arrhythmogenic Cardiomyopathy Cohort. Int. J. Mol. Sci. 2020, 21, 1536.
- Khudiakov, A.A.; Panshin, D.D.; Fomicheva, Y.V.; Knyazeva, A.A.; Simonova, K.A.; Lebedev, D.S.; Mikhaylov, E.N.; Kostareva, A.A. Different Expressions of Pericardial Fluid MicroRNAs in Patients with Arrhythmogenic Right Ventricular Cardiomyopathy and Ischemic Heart Disease Undergoing Ventricular Tachycardia Ablation. Front. Cardiovasc. Med. 2021, 8, 647812.
- Desantis, V.; Potenza, M.A.; Sgarra, L.; Nacci, C.; Scaringella, A.; Cicco, S.; Solimando, A.G.; Vacca, A.; Montagnani, M. microRNAs as Biomarkers of Endothelial Dysfunction and Therapeutic Target in the Pathogenesis of Atrial Fibrillation. Int. J. Mol. Sci. 2023, 24, 5307.
- Hao, H.; Dai, C.; Han, X.; Li, Y. A novel therapeutic strategy for alleviating atrial remodeling by targeting exosomal miRNAs in atrial fibrillation. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119365.
- Tan, A.Y.; Zimetbaum, P. Atrial fibrillation and atrial fibrosis. J. Cardiovasc. Pharmacol. 2011, 57, 625–629.
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular Mechanisms Driving mRNA Degradation by m6A Modification. Trends Genet. 2020, 36, 177–188.
- Winkler, G.S. The mammalian anti-proliferative BTG/Tob protein family. J. Cell. Physiol. 2010, 222, 66–72.
- Matsuda, S.; Rouault, J.; Magaud, J.; Berthet, C. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 2001, 497, 67–72.
- Tirone, F. The gene PC3TIS21/BTG2, prototype member of the PC3/BTG/TOB family: Regulator in control of cell growth, differentiation, and DNA repair? J. Cell. Physiol. 2001, 187, 155–165.
- Ezzeddine, N.; Chang, T.-C.; Zhu, W.; Yamashita, A.; Chen, C.-Y.A.; Zhong, Z.; Yamashita, Y.; Zheng, D.; Shyu, A.-B. Human TOB, an Antiproliferative Transcription Factor, Is a Poly(A)-Binding Protein-Dependent Positive Regulator of Cytoplasmic mRNA Deadenylation. Mol. Cell. Biol. 2007, 27, 7791–7801.
- Ikeda, Y.; Taniguchi, K.; Nagase, N.; Tsuji, A.; Kitagishi, Y.; Matsuda, S. Reactive oxygen species may influence on the crossroads of stemness, senescence, and carcinogenesis in a cell via the roles of APRO family proteins. Explor. Med. 2021, 2, 443–454.
- Ikeda, Y.; Taniguchi, K.; Sawamura, H.; Yoshikawa, S.; Tsuji, A.; Matsuda, S. Presumed Roles of APRO Family Proteins in Cancer Invasiveness. Cancers 2022, 14, 4931.
- Zhang, J.; Dong, W. Expression of B Cell Translocation Gene 1 Protein in Colon Carcinoma and its Clinical Significance. Recent Pat. Anti-Cancer Drug Discov. 2020, 15, 78–85.
- Zhao, S.; Xue, H.; Hao, C.L.; Jiang, H.M.; Zheng, H.C. BTG1 Overexpression Might Promote Invasion and Metastasis of Colorectal Cancer via Decreasing Adhesion and Inducing Epithelial-Mesenchymal Transition. Front. Oncol. 2020, 10, 598192.
- Jung, Y.Y.; Sung, J.Y.; Kim, J.Y.; Kim, H.S. Down-regulation of B-Cell Translocation Gene 1 by Promoter Methylation in Colorectal Carcinoma. Anticancer Res. 2018, 38, 691–697.
- Ishida, Y.; Kawakami, H.; Kitajima, H.; Nishiyama, A.; Sasai, Y.; Inoue, H.; Muguruma, K. Vulnerability of Purkinje Cells Generated from Spinocerebellar Ataxia Type 6 Patient-Derived iPSCs. Cell Rep. 2016, 17, 1482–1490.
- Luan, S.H.; Yang, Y.Q.; Ye, M.P.; Liu, H.; Rao, Q.F.; Kong, J.L.; Wu, F.R. ASIC1a promotes hepatic stellate cell activation through the exosomal miR-301a-3p/BTG1 pathway. Int. J. Biol. Macromol. 2022, 211, 128–139.
- Hwang, S.S.; Lim, J.; Yu, Z.; Kong, P.; Sefik, E.; Xu, H.; Harman, C.C.D.; Kim, L.K.; Lee, G.R.; Li, H.B.; et al. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science 2020, 367, 1255–1260.
- Li, B.N.; Tang, Q.D.; Tan, Y.L.; Yan, L.; Sun, L.; Guo, W.B.; Qian, M.Y.; Chen, A.; Luo, Y.J.; Zheng, Z.X.; et al. Key Regulatory Differentially Expressed Genes in the Blood of Atrial Septal Defect Children Treated with Occlusion Devices. Front. Genet. 2021, 12, 790426.
- Tzachanis, D.; Boussiotis, V.A. TOB, a member of the APRO family, regulates immunological quiescence and tumor suppression. Cell Cycle 2009, 8, 1019–1025.
- Lee, H.S.; Kundu, J.; Kim, R.N.; Shin, Y.K. Transducer of ERBB2.1 (TOB1) as a Tumor Suppressor: A Mechanistic Perspective. Int. J. Mol. Sci. 2015, 16, 29815–29828.




