Genome reduction is a top-down approach to achieve the minimal genetic information essential for a living cell. Exploring the minimal genetic requirements for cells to maintain free living is an exciting topic in biology. Multiple approaches are employed to address the question of the minimal genome. In addition to constructing the synthetic genome in the test tube, reducing the size of the wild-type genome is a practical approach for obtaining the essential genomic sequence for living cells. The well-studied Escherichia coli has been used as a model organism for genome reduction owing to its fast growth and easy manipulation.
1. Introduction
The genome is the blueprint of life, including all the living organism’s information. The abundance of genetic information a genome contains is nearly determined by the number of base pairs encoded on the genome, both the genes and the non-coding regions. The quantitative evaluation of the genetic abundance is known as the genome size, which largely varies among organisms. In general, the more complex the structure and ecology of an organism, the larger its genome size. For example, a typical virus has a tiny genome size of about 1~30 kbp
[1], while a human cell has a genome of 6.4 Gb as diploid, and one of the ferns,
Paris japonica, has a giant genome of 597 Gb as octaploid
[2][3]. It is known that the genome size range in archaea and bacteria is between 100 kbp and 16 Mb, which can be linked to the microorganism’s ecosystem type (aquatic, host-associated, or terrestrial)
[4]. The genome size roughly represents the abundance of the genetic information required for its holder cell.
Living cells are maintained by transcribing genes encoded in the genome into mRNA, followed by their translation into proteins, which shape cells and catalyse various chemical reactions. The relationship between the genome and the phenotype, the growth fitness, has often been investigated. In addition, genomes have reached their current form through mutations and horizontal gene transfer during evolution; thus, it is crucial to examine genomes to unravel the evolution of organisms. The genome size is the consequence of evolution, which either increases or decreases the genomic sequences. What the minimal genetic information needed is for life on Earth remains an open question.
2. Genome Reduction
Escherichia coli (
E. coli) has been widely used in synthetic biology due to its fast growth in rich media
[5] and high transformation and plasmid integration efficiency
[6]. Nevertheless, the essential and substantial genetic requirements of
E. coli are not fully understood. Although
E. coli has been well studied in genetics and bioengineering, and its first genome sequence was determined approximately 25 years ago
[7], the molecular and physiological functions across the genome have not been fully clarified. Most
E. coli genomes range from 3.8 Mb to more than 6 Mb, and the average is around 5 Mb.
E. coli K-12 MG1655, one of the most well-known
E. coli strains, has about 4.6 Mb and contains approximately 4400 genes, highly interacting with each other and shaping various biological networks. Gene regulatory network (GRN) and transcriptional regulatory network (TRN) were constructed to know the regulatory relationship of genes and often reconstructed to improve the prediction of gene expression
[8][9][10][11]. Connecting GRN to the metabolic network, which is consisted of metabolites, was implemented to understand the cell system
[12]. It is also indicated current GRN may not be able to predict gene expression
[13]. A number of genes still have unknown functions, and understanding the entire cell as a dynamic system is challenging due to the complexity of genetic and metabolic networks. In other words, the aim of
E. coli genomics is to reveal a regulatory network with thousands of elements, but the genome is far too complex for this purpose.
To discover genetic essentiality, reducing the genetic elements has been challenged to a large extent. Significant efforts have been made to construct a minimal genome containing the essential genes for growing under the defined conditions to provide the genetically simplest model. There are two main approaches to constructing a minimal genome: bottom-up and top-down. The bottom-up approach is represented by landmark works that chemically synthesize a genome containing only essential genes and transfer it to the cytoplasm
[14][15][16]. The top-down approach involves genetic deletions by removing redundant DNA sequences from the wild-type (full-length)
E. coli genomes, known as genome reduction (
Figure 1). Such genetic deletion approaches were employed to a great extent to identify the minimal genetic requirement for a bacterial cell.
Figure 1. Schematic drawing of genome reduction. A reduced genome is constructed by removing redundant genomic sequences from the parent genome (wild-type genome).
So far, the reduced genomes were mainly constructed with the
E. coli strains of MG1655 and W3110, two representative full-length genomes of approximately 4.6 Mb. Multiple deletions of genomic sequences were commonly performed with the traditional genetic methodologies, e.g., λ Red recombinase and P1 transduction, which were conducted many years ago
[17][18]. After the CRISPR/Cas9 system was developed and used widely
[19][20], the random deletion method combining CRISPR/Cas9 and transposon also became available to reduce the genome
[21]. Comparative genomics and computational approaches are also available for investigating the genome range of reducible
[22]. Various genome-reduced
E. coli strains have been successfully constructed in laboratories
[23][24][25][26], and those of extensive deletions (i.e., more than 10% of the parent wild-type genomes are absent) are summarized (
Table 1).
Table 1. Representative genome-reduced
E. coli strains. The wide-type strains used as the parent genomes for multiple deletions, the resultant reduced genomes, and their growth fitness in different growth media are summarized according to the previous report
[27].
Parent
Genome |
Strain Name |
Genome Size (Mb) |
Reduced Ratio |
Growth
Medium |
Growth
Fitness |
W3110
(4.66 Mb) |
MGF-01 (N28) |
3.6 |
22% |
minimal |
decreased |
| minimal, amino acids |
decreased |
| rich |
decreased |
| DGF-298 |
3.0 |
36% |
rich |
increased |
MG1655
(4.64 Mb) |
MDS42 |
4.0 |
14% |
minimal |
equivalent |
| rich |
equivalent |
| minimal, amino acids |
decreased |
| MDS69 |
3.7 |
20% |
rich |
decreased |
| Δ16 |
3.3 |
30% |
minimal |
decreased |
| MS56 |
3.6 |
23% |
minimal |
increased, decreased |
| rich |
equivalent, decreased |
The minimal genomes were considered beneficial for material production and genetic recombination. For instance, the successful downstream applications of the reduced genomes summarized in
Table 1 were reported. The genome-reduced
E. coli strain MGF-01 showed increased L-threonine production compared to the wild-type strain W3110 carrying the full-length genome
[28][29]. The reduced genome MDS42 was constructed by removing the IS elements, which made it highly useful for genetic recombination without IS-mediated mutagenesis. The efficiency of DNA transformation of MDS42 was over 180-fold higher than that of its parent strain MG1655 and was equal to or higher than that of commercially available competent cells
[24]. The recombinant Isoamylase derived from
Thermobifida fusca has been successfully produced using this strain
[30], and the modified strain MDS-205 has improved L-threonine production
[31]. Genome reduction of
Bacillus is also a well-known application of the bacterial minimal genome. As a representative example,
Bacillus subtilis PG10 has about 36% of its genome deleted, which is industrially applicable with its enhanced protein secretion capacity
[32][33]. Additionally, the genome-reduced
Bacillus amyloliquefaciens strain GR167, lacking approximately 4.18% of the genomic sequence of the parent strain LL3, was constructed and successfully produced 311.35 mg/L of surfactin, a biosurfactant
[34].
Deleting the redundant DNA sequences, which are supposed to be unnecessary for living cells, causes differentiated changes in growth fitness. In particular, the reduced genomes of large deletions showed a decrease in growth rate compared to their parental strains under nutritionally poor media (
Table 1). The high-throughput growth assay of the reduced genomes found the growth rates were correlated to the deleted length of genomic sequences
[35]. That is, the genome’s fitness is somehow connected with its size. In addition, genome reduction is likely to cause an increase in spontaneous mutation rate in a deletion size-correlated manner
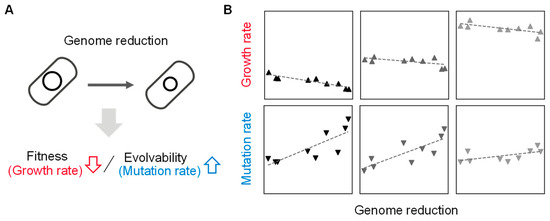
[36][37]. These studies demonstrated that genome size, growth rate, and mutation rate were quantitatively associated, suggesting that the abundance of genetic information, i.e., genome, is highly coordinated with the fitness and evolvability of the living cells (
Figure 2). Even genomic sequences that are nonessential for growing in normal conditions, e.g., rich media, play a role in accelerating cell growth. Genome reduction without growth decrease is crucial for addressing the question of what the minimal genome is. Besides the synthetic approach, i.e., conventional genetic construction, the evolutionary and environmental approaches should be considered to acquire extensively reduced genomes.
Figure 2. Coordination of genome size to growth and mutation rates. (
A) Contribution of genome reduction to fitness and evolvability. The changes in growth and mutation rates caused by the genome reduction are indicated with arrows. (
B) Relative values of genome, growth, and mutation. Gray gradation indicates the variation in growth media. The scatter plots are newly made using previously reported data
[37][38]. The panels from left to right represent the nutritional richness of culture media from poor to rich.
3. Evolutionary Approaches for Reduced Genome
In wild nature, free-living bacteria were found to show high growth fitness despite holding small genomes, such as
Pelagibacter ubique, an abundant living organism in seawater. It can grow quickly, even with a considerably small genome size of only 20% of that of
E. coli (e.g., 1 Mb compared to 5 Mb)
[39][40]. Intriguingly, the
Pelagibacter ubique genome contains few transposons or pseudogenes, indicating the reduced genome could have high fitness. It was considered that the living cells had acquired their current fitness through natural evolution. Evolution can be a powerful tool in searching for an essential set of genes to gain improved fitness in the habit (i.e., living environment). Experimental evolution mimics nature evolution under well-controlled laboratory conditions
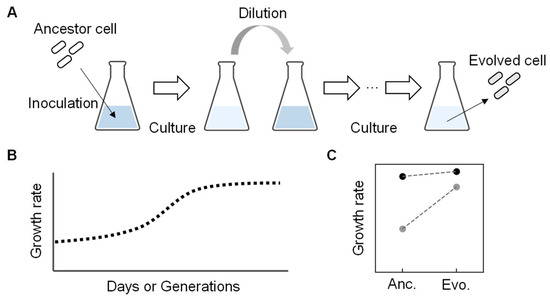
[41][42]. It is generally conducted through the serial transfer of the bacterial populations to select derivatives with improved fitness. The serial transfer is repeated inoculation and dilution of a portion of the bacterial population grown to the early exponential growth phase into a fresh medium with the same composition (
Figure 3A). The end-point (evolved) population finally achieves higher fitness than the initial population (ancestor) (
Figure 3B), so it is also called adaptive laboratory evolution (ALE)
[43]. The improved fitness of evolved bacterial populations was usually accompanied by the accumulation of genome mutations during evolution. Interestingly, when the experimental evolution of an identical ancestor was performed with multiple lineages under the same condition in parallel, the genome mutations found in the final evolved populations often differed among the lineages
[44][45]. The genomic location of the mutations and the timing of the mutations fixing on the genome varied among the parallelly evolved lineages
[46][47]. It indicates that the experimental evolution provides multiple trajectories for the ancestral genome to acquire somehow finetuned genomic sequences promising growth fitness. So far, intensive studies have demonstrated that experimental evolution is a powerful approach to selecting a bacterial population (genotype) with an increased growth rate (relative fitness)
[42][48].
Figure 3. An overview of experimental evolution. (
A) Experimental evolution. Repeated culture and dilution are performed with the ancestor to acquire the evolved genome with an improved growth rate. (
B) Temporal changes in growth rate during experimental evolution. (
C) Growth rate changes between the ancestor and evolved
E. coli cells. Dark and light circles represent the full-length and reduced genomes, respectively. The previously reported data
[49] were used to make the graph.
Experimental evolution of reduced genomes has been reported with different
E. coli strains. The reduced genomes somehow show differentiated evolvability compared to the wild-type genomes. As the deleted size of the genome correlated with the increased mutation rate (i.e., improved evolvability)
[37], the removal of IS elements or disabling other mutation-generating pathways of the host could result in reduced evolvability
[50][51]. Although, the adaptive evolution to the new environmental conditions could be successfully achieved independent of their full-length parent genomes, e.g., MG1655 and W3110
[45][49][52]. Four reduced genomes lacking 200~1000 kb DNA sequences were evolved for ~1000 generations, and the growth rates of these reduced genomes were raised to an equivalent level to that of the wild-type (full-length) genome
[49]. These results show that the decreased growth rate caused by the deletion of a large gene set can be complemented by introducing a few small mutations, e.g., single-nucleotide substitutions, without inserting additional genes.
In addition, the changes in mutation rates were coordinated with the increase in growth rates
[37][38], revealing that the experimental evolution compensated for the genome reduction-mediated changes in growth and mutate rates. Note that such coordination might be stringently related to transcriptome reorganization. Although the gene expression patterns were significantly disturbed by genome reduction and experimental evolution, the chromosomal periodicity and negative epistasis (i.e., canceling effect) of the transcriptomes were observed
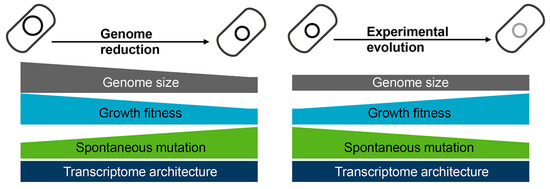
[53]. In a word, experimental evolution rescued the growth and mutation rates disturbed by genome reduction (
Figure 4). Intriguingly, the changes in growth fitness caused by either genome reduction or experimental evolution were dependent on the deletion size
[37][49], indicating the abundance of genetic information but not the specific gene function participate in the coordinated relationships. Although the underlying mechanisms are unclear, maintaining the homeostatic chromosomal architecture must be crucial for optimizing the growth rate of cells with reduced genomes.
Figure 4. Changes caused by genome reduction and rescued via experimental evolution. Genome reduction causes decreased growth fitness and increased mutation rate (left panel), which is restored via experimental evolution (right panel). The transcriptome architecture maintains homeostasis regardless of genome reduction or experimental evolution.