You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Busheng Xue | -- | 1810 | 2023-08-17 09:39:07 | | | |
| 2 | Catherine Yang | -4 word(s) | 1806 | 2023-08-17 09:42:13 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Gong, H.; Xue, B.; Ru, J.; Pei, G.; Li, Y. Targeted Therapy for EWS-FLI1 in Ewing Sarcoma. Encyclopedia. Available online: https://encyclopedia.pub/entry/48154 (accessed on 29 December 2025).
Gong H, Xue B, Ru J, Pei G, Li Y. Targeted Therapy for EWS-FLI1 in Ewing Sarcoma. Encyclopedia. Available at: https://encyclopedia.pub/entry/48154. Accessed December 29, 2025.
Gong, Helong, Busheng Xue, Jinlong Ru, Guoqing Pei, Yan Li. "Targeted Therapy for EWS-FLI1 in Ewing Sarcoma" Encyclopedia, https://encyclopedia.pub/entry/48154 (accessed December 29, 2025).
Gong, H., Xue, B., Ru, J., Pei, G., & Li, Y. (2023, August 17). Targeted Therapy for EWS-FLI1 in Ewing Sarcoma. In Encyclopedia. https://encyclopedia.pub/entry/48154
Gong, Helong, et al. "Targeted Therapy for EWS-FLI1 in Ewing Sarcoma." Encyclopedia. Web. 17 August, 2023.
Copy Citation
Ewing sarcoma (EwS) is a highly aggressive and metastatic cancer in children and adolescents. Canonical therapy mainly comprises the combination of intensive chemotherapy, radiation, and local surgery, which give rise to acute and chronic adverse effects. Drugs targeting EwS without side effects are in urgent demand. Genetically, EwS is characterized by chromosomal translocations with a low mutation burden. As a result, the chimeric protein EWS-ETS, mainly EWS-FLI1(85%), is critical for the malignancy of EwS. EWS-FLI1 directly binds to GGAA microsatellites in enhancers and promotors of the target genes and recruits multiple transcription factors or epigenetic regulators to reprogramme the epigenome.
ewing sarcoma
EWS-FLI1
protein complex
targeted therapy
1. Introduction
Ewing sarcoma (EwS) is a poorly differentiated malignancy that mainly arises in bone and soft tissue and is more prevalent among those in the second decade of life [1]. Extraosseous EwS is more common in adults [2]. The cellular origin of EwS remains controversial, although it is speculated that it arises from neuroectodermal cells or primitive mesenchymal stem cells (MSC) [3][4][5]. Despite considerable improvements in overall survival achieved using a multimodal approach, including intensive chemotherapy for localized disease [6], the prognosis of patients who develop metastatic disease or relapse remains dismal [7][8]. Approximately 20–25% of patients are diagnosed with early micro-dissemination [6][9].
2. Involvement of EWS-FLI1 in Transcription, Epigenetic Reprogramming, and Alternative Splicing in EwS
2.1. EWS-FLI1 in Transcription and Epigenetic Reprogramming
EWS-FLI1 is an aberrant transcription factor that drives cellular transformation by rewiring the epigenome to induce a large number of oncogenes. The N-terminus of EWSR1 contains a prion-like domain, characterized by an intrinsically disordered structure and low complexity. This domain has phase transition properties and manipulates multiple proteins involved in epigenome reprogramming and epigenetic alterations [10][11][12][13][14][15][16][17]. In addition to the canonical ETS-binding sites, EWS-FLI1 binds to DNA sequences at the GGAA/T core motif [18][19][20] via a conserved ETS domain. It regulates multiple proteins through its prion-like domain to tumor-specific enhancers and promotors, recruiting acetyltransferases and establishing de novo enhancers by generating H3K27ac, thus opening the chromosomal architecture, which contributes to the activation of target genes [10][11][18][20]. The EWS-FLI1 protein complex includes RNA polymerase II [21][22], the core subunit hsRBP7 (human RNA polymerase II) [23][24], E2F3 [25][26], EWSR1 [27], CBP/p300 [28], WDR5, ASH2, MLL [11], and the BAF complex (mammalian SWI/SNF complex) [10][29]. The threshold of GGAA motifs optimal for maximal expression is 20–26 [30], which differs from that in wild-type FLI1. Super-enhancer-associated MEIS1 and RING1B also contribute to the chromatin reprogramming through co-localization with EWS-FLI1 at the active enhancers to drive the malignancy of EwS [31][32]. This specific coupling results in the activation of many genes (Figure 1), such as NKX2.2 [33], NROB1 [34][35][36], IGF1R [37], BCL11B [38], EZH2 [17], VRK1 [11], GLI1 [39], PTPL1 [40], PPPR1A [41], ERG2 [42], GSTM4 [43], PAX7 [44], CHM1 [45], REST [46], PHF19 [13], STEAP1 [47][48], SLFN11(Schlafen 11) [49], HDAC3 [50], TNC [51], APCDD1 [31], IL1RAP [52][53], MYC [54], and PRC1 (protein regulator of cytokinesis 1) [55].
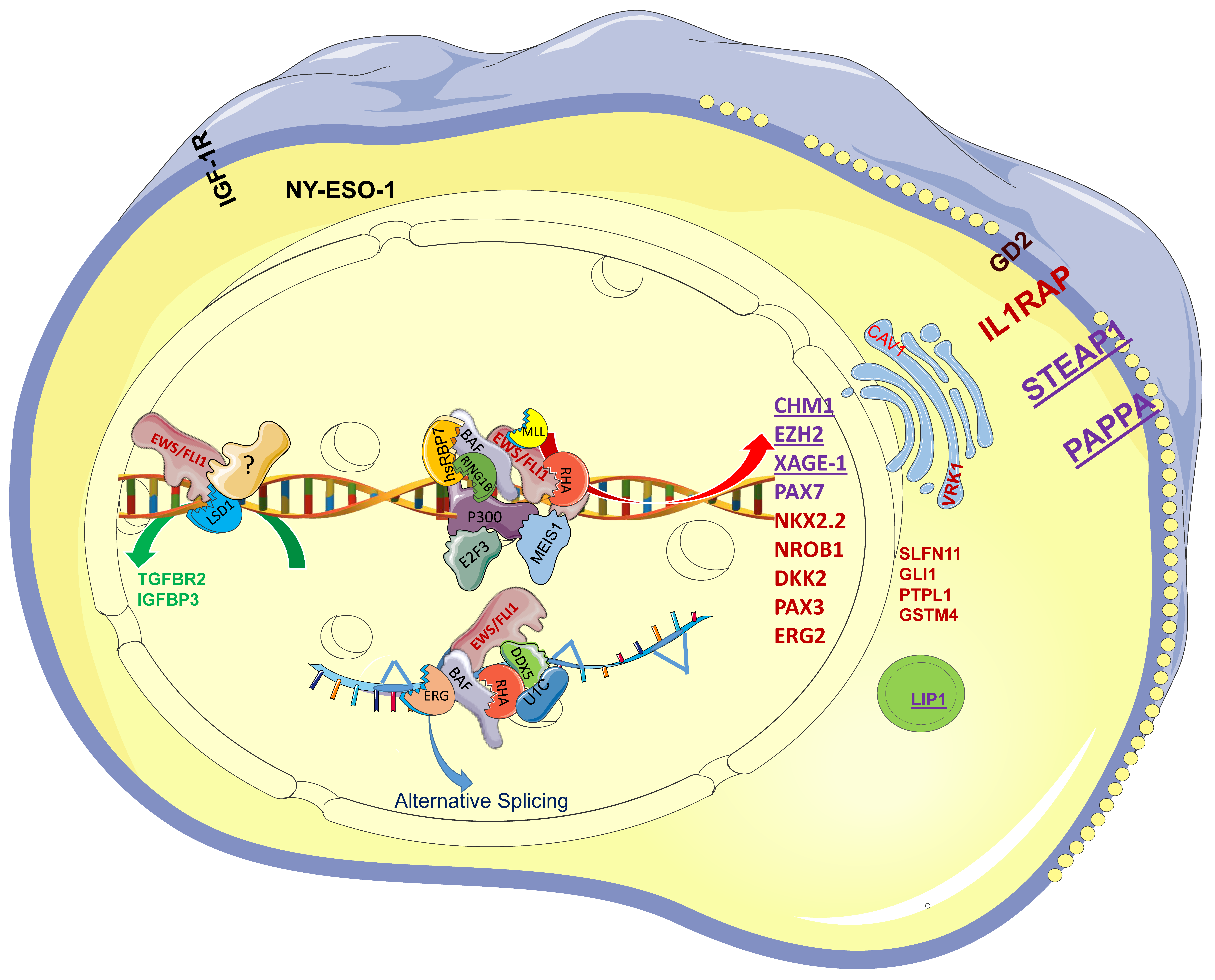
Figure 1. The EWS-FLI1 protein complex drives the specific transcription profile of EwS. EWS-FLI1 recruits E2F3, hsRBP7, BAF, RING1B, RHA, P300, and MEIS1, among others, to GGAA repeats and further activates CHM1, EZH2, PAX7, NKX2.2, NROB1, and STEAP1, among others. EWS-FLI1 functions as a protein complex with ERG, BAF, RHA, DDX5, and U1C to drive alternative splicing. Among the genes, the purple ones, such as CHM1, could serve as TCR-based immunotherapy targets; the red ones, such as NKX2-2, serve as diagnostic markers in clinic diagnosis. EWS/FLI1 recruits LSD1 and unknown transcription factors (?) to repress TGFBR1 and IGFBP3, which still needs further research.
Among the direct targets of EWS-FLI1, NKX2-2 mediates oncogenic transformation via transcriptional repression and is necessary and sufficient for the oncogenic phenotype of EwS [56]. Further work demonstrates that NKX2-2, KLF15, and TCF4 occupy similar super-enhancers and promoters, forming an inter-connected auto-regulatory loop and occupying 77.2% of promoters and 55.6% of enhancers shared with EWS-FLI1 [57], such as STEAP1 [58]; this kind of coordinate regulation drives the proliferation of EwS. NROB1 directly interacts with EWS-FLI1 to modulate multiple gene expressions and mediate the oncogenic phenotype of EwS [59]. SLFN11 is a putative DNA/RNA helicase that recruits to the stressed replication fork and irreversibly triggers replication block and cell death. Overexpression of SLFN11 is associated with resistance to topoisomerase I inhibitors and poly (ADP-ribose) polymerase (PARP) inhibitor combinations [49][60]. STEAP1 and IL1RAP are vital for the redox homeostasis of EwS [47][52]. APCDD1, PHF19, GSTM4, and PTPL1 are genes that are involved in the proliferation of EwS.
EWS-FLI1 is also involved in transcriptional repression of tumor suppressors such as IGFBP3 [61] and PHLDA1 [35] to drive oncogenic transformation [12][62]. The nucleosome remodeling and deacetylase (NuRD) complex is a typical ATP-dependent chromatin remodeling complex [63] that plays a critical role in transcription and determines differentiation and development [64]. EWS-FLI1 recruits the NuRD-LSD1 complex to repress LOX and TGFBR2 [62][65]. It also affects the transcriptional activation of AP-1 [14] and MRTFB [66] and binds to the promotor of FOXO1 to repress its expression, thereby increasing tumor growth [67]. EWS-FLI1 promotes the phosphorylation of cyclin-dependent kinase-2 and AKT to inhibit the activity of FOXO1, thus rewiring transcriptional repression [67]. EWS-FLI1 is also involved in the regulation of microRNAs (miRNAs) [68]. It downregulates miRNA-145 to initiate mesenchymal stem-cell reprogramming toward EwS stem cells [69] and represses miR-708, which induces the overexpression of EYA3 and contributes to the chemoresistance to etoposide and doxorubicin [70].
The histone methyltransferase EZH2 exhibits silencing activity via methylation of H3K27 [71]. In EwS, EWS-FLI1 upregulates EZH2 expression by interacting with the EZH2 promoter, thereby promoting tumor growth/metastasis and blocking endothelial/neuro-ectodermal differentiation [17].
MiR-34a inhibits the proliferation and increases the sensitivity of EwS to doxorubicin and vincristine and is a strong predictor of a favorable prognosis in EwS [72]. However, the exact mechanism underlying its downregulation remains elusive. Exportin 5 (XPO5), which mediates the nuclear export of pre-miRNAs and short hairpin RNAs [73][74][75], interacts with EWS-FLI1 based on mass spectrometry [76]. XPO5 is highly expressed in various cancers including EwS. Furthermore, the phosphorylation of XPO5 alters the nucleus and cytoplasm shift [77]. Investigating XPO5 and its relationship with EWS-FLI1 may offer new insights into the therapy of EwS. Post-translational modifications of EWS-FLI1 modulate its transcriptional activity. Phosphorylation and O-GlcNAcylation of the N-terminus of EWSR1 [78][79][80], as well as acetylation of the C-terminal FLI1 domain by PCAF (KAT2B, lysine acetyltransferase 2B), enhance the transcriptional activity of EWS-FLI1 [81]. However, PCAF expression is lower in EwS tissues, which is a common feature of cancer.
2.2. EWS-FLI1 in Alternative Splicing
Pre-mRNA splicing is critical for gene expression, and most protein-encoding transcripts are alternatively spliced to provide diverse functions [82][83]. The N-terminus of EWSR1 interacts with the hyperphosphorylated RNA polymerase II and recruits serine-arginine (SR) through its C-terminus. After chromosome translocation, the C-terminus of wild-type EWSR1 is replaced by FLI1, which hinders the recruitment of SR-splicing factors and interferes with mRNA splicing [21], thus demonstrating the negative property of this chimeric protein [84]. This leads to comprehensive alternative splicing of numerous genes. Meanwhile, EWS-FLI1 interacts with the splicing components (snRNP) U1C and SF1 to modulate pre-mRNA splicing [85]. It also recruits the BAF complex to drive the alternative splicing of ARID1A and the preferential splicing of ARID1A-L, which is necessary for tumor growth [86]. Work by Selvanathan [76] demonstrates that EWS-FLI1 is involved in the alternative splicing of CLK1, CASP3, PPFIBP1, and TERT, which potentially regulate the oncogenesis of EwS.
3. The Regulation of EWS-FLI1
Transcription and post-transcriptional modifications are involved in the regulation of expression and activity of EWS-FLI1. Although the transcriptional regulation of EWS-FLI1 remains elusive, the BRD4 inhibitor JQ1 suppresses this activity [13][15][87]. HDAC6 deacetylates specificity protein 1 (SP1), thereby inhibiting the recruitment of the SP1/P300 complex to the promoters of EWSR1 and EWS-FLI1 and downregulating EWS-FLI1 [88]. MiR-145 and let-7 repress EWS-FLI1 by targeting its mRNA [69][89][90][91] and inhibit the proliferation of EwS. The RNA-binding protein LIN28B interacts with EWS-FLI1 transcripts to maintain the stability and ensure the expression of EWS-FLI1 to enhance the tumorigenicity of the self-renewal of EwS [90]. At the post-transcriptional level, EWS-FLI1 degradation is proteasome dependent, and the protein has a half-life of 2–4 h [92]. This process can be protected by the action of ubiquitin-specific protease 19 (USP19) at the N-terminus [93] and accelerated by tripartite-motif-containing 8 (TRIM8) at K334 [94]; however, USP19 is expressed at low levels in EwS. Casein kinase 1 (CK1)-mediated phosphorylation of the VTSSS degron in the FLI1 domain activates speckle-type POZ protein (SPOP) activity, which degrades EWS-FLI1. In contrast, OTU-domain-containing protein 7A (OTUD7A) participates in the deubiquitination of the C-terminus and stabilizes EWS-FLI1 [95]. The inhibitor of chromosomal maintenance 1 (CRM1 and XPO1), KPT-330 [96], and IFN-γ [97] suppress expression of EWS-FLI1 at the protein level. FOXM1, a downstream factor of EWS-FLI1, upregulates its expression [98]. Cytosine arabinoside (ARA-C) downregulates EWS-FLI1 at the protein level and inhibits tumor growth [99]; however, it shows hematologic toxicity and minimal activity in patients [100].
STAG2 (stromal antigen 2) is a core subunit of the cohesion complex and is frequently mutated in multiple cancers [101] including EwS [102][103]. Mutation of STAG2 in EwS is associated with poor outcomes by improving metastasis [102]. Mechanically, in addition to the disruption of PRC2-mediated regulation of gene expression in EwS [104], the inactivation of STAG2 strongly altered CTCF-anchored loop extrusion and decreases promotor-enhancer interactions. As a result, the cis-mediated EWS-FLI1 activity at GGAA microsatellite neo-enhancers is downregulated and the cells are enhanced in their migration and invasion properties [104][105].
Unlike STAG2, there was no evidence showing mutations of the ETS transcription factor ETV6 in EwS [106]. ETV6 does not change the expression of EWS-FLI1 but co-occupy loci genome wide at the short consecutive GGAA repeats and constrains the transcriptional activity of EWS-FLI1 [107][108]. Upon inactivating ETV6, EWS-FLI1 overtakes and activates these cis-elements to promote mesenchymal differentiation by upregulating the expression of SOX11 [108].
4. CAR-T Therapy
Unlike TCR-based T-cell therapy, which is limited to specific HLA restriction and deficient HLA expression in EwS [109] because of the presence of myeloid-derived suppressor cells, F2 fibrocytes, and M2-like macrophages in the microenvironment [110], CAR-engineered T-cell therapy can target specific cell-surface antigens in tumors, independent of HLA. VEGFR2 is a potential target for CAR-T-cell therapy in EwS [111]. In addition to TCR-T-cells, CAR-T-cells [112] can target STEAP1, which is involved in the malignant phenotype of EwS [47].
CAR-T targeting GPR64, ROR1, and IGF1R, which are highly expressed in EwS [113][114], leads to a selective killing of EwS in vivo [115]. LINGO1, which is highly expressed in EwS [116], is a direct target of EWS-FLI1. EZH2 inhibition by GSK-126 induces GD2 surface expression in EwS [117], and the combination of CAR-T therapy targeting GD2 and EZH2 inhibitors have synthetic cytotoxic in the treatment of EwS; this kind of combination provides new options for the clinical application. IL1RAP, a direct target of EWS-FLI1, is highly expressed in EwS, but minimally expressed in normal tissues, which makes it a promising surface target for EwS [52] and a potential candidate for advanced CAR-T therapy. ICAM-1 can promote tumor cell/T-cell interaction and T-cell activation, and the knockdown of EWS-FLI1 upregulates ICAM-1 expression and leads to the upregulation of PD-L1 and PD-L2, both proteins that inhibit the activity of T-cells [97]. Blocking PD-1 with a checkpoint inhibitor could increase the T-cell-mediated killing of EwS cells with low expression of EWS-FLI1.
References
- Grunewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; de Alava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Primers 2018, 4, 5.
- Jahanseir, K.; Folpe, A.L.; Graham, R.P.; Giannini, C.; Robinson, S.I.; Sukov, W.; Fritchie, K. Ewing Sarcoma in Older Adults: A Clinicopathologic Study of 50 Cases Occurring in Patients Aged >/=40 Years, With Emphasis on Histologic Mimics. Int. J. Surg. Pathol. 2020, 28, 352–360.
- Riggi, N.; Suva, M.L.; Stamenkovic, I. Ewing’s sarcoma origin: From duel to duality. Expert Rev. Anticancer Ther. 2009, 9, 1025–1030.
- Tirode, F.; Laud-Duval, K.; Prieur, A.; Delorme, B.; Charbord, P.; Delattre, O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 2007, 11, 421–429.
- Kovar, H. Blocking the road, stopping the engine or killing the driver? Advances in targeting EWS/FLI-1 fusion in Ewing sarcoma as novel therapy. Expert Opin. Ther. Targets 2014, 18, 1315–1328.
- Gaspar, N.; Hawkins, D.S.; Dirksen, U.; Lewis, I.J.; Ferrari, S.; Le Deley, M.C.; Kovar, H.; Grimer, R.; Whelan, J.; Claude, L.; et al. Ewing Sarcoma: Current Management and Future Approaches Through Collaboration. J. Clin. Oncol. 2015, 33, 3036–3046.
- Haeusler, J.; Ranft, A.; Boelling, T.; Gosheger, G.; Braun-Munzinger, G.; Vieth, V.; Burdach, S.; van den Berg, H.; Juergens, H.; Dirksen, U. The value of local treatment in patients with primary, disseminated, multifocal Ewing sarcoma (PDMES). Cancer 2010, 116, 443–450.
- Zollner, S.K.; Amatruda, J.F.; Bauer, S.; Collaud, S.; de Alava, E.; DuBois, S.G.; Hardes, J.; Hartmann, W.; Kovar, H.; Metzler, M.; et al. Ewing Sarcoma-Diagnosis, Treatment, Clinical Challenges and Future Perspectives. J. Clin. Med. 2021, 10, 1685.
- Ladenstein, R.; Potschger, U.; Le Deley, M.C.; Whelan, J.; Paulussen, M.; Oberlin, O.; van den Berg, H.; Dirksen, U.; Hjorth, L.; Michon, J.; et al. Primary disseminated multifocal Ewing sarcoma: Results of the Euro-EWING 99 trial. J. Clin. Oncol. 2010, 28, 3284–3291.
- Boulay, G.; Sandoval, G.J.; Riggi, N.; Iyer, S.; Buisson, R.; Naigles, B.; Awad, M.E.; Rengarajan, S.; Volorio, A.; McBride, M.J.; et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171, 163–178.e119.
- Riggi, N.; Knoechel, B.; Gillespie, S.M.; Rheinbay, E.; Boulay, G.; Suva, M.L.; Rossetti, N.E.; Boonseng, W.E.; Oksuz, O.; Cook, E.B.; et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014, 26, 668–681.
- Sankar, S.; Theisen, E.R.; Bearss, J.; Mulvihill, T.; Hoffman, L.M.; Sorna, V.; Beckerle, M.C.; Sharma, S.; Lessnick, S.L. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clin. Cancer Res. 2014, 20, 4584–4597.
- Gollavilli, P.N.; Pawar, A.; Wilder-Romans, K.; Natesan, R.; Engelke, C.G.; Dommeti, V.L.; Krishnamurthy, P.M.; Nallasivam, A.; Apel, I.J.; Xu, T.; et al. EWS/ETS-Driven Ewing Sarcoma Requires BET Bromodomain Proteins. Cancer Res. 2018, 78, 4760–4773.
- Tomazou, E.M.; Sheffield, N.C.; Schmidl, C.; Schuster, M.; Schonegger, A.; Datlinger, P.; Kubicek, S.; Bock, C.; Kovar, H. Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS-FLI1. Cell Rep. 2015, 10, 1082–1095.
- Richter, G.H.S.; Hensel, T.; Schmidt, O.; Saratov, V.; von Heyking, K.; Becker-Dettling, F.; Prexler, C.; Yen, H.Y.; Steiger, K.; Fulda, S.; et al. Combined Inhibition of Epigenetic Readers and Transcription Initiation Targets the EWS-ETS Transcriptional Program in Ewing Sarcoma. Cancers 2020, 12, 304.
- Sheffield, N.C.; Pierron, G.; Klughammer, J.; Datlinger, P.; Schonegger, A.; Schuster, M.; Hadler, J.; Surdez, D.; Guillemot, D.; Lapouble, E.; et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat. Med. 2017, 23, 386–395.
- Richter, G.H.; Plehm, S.; Fasan, A.; Rossler, S.; Unland, R.; Bennani-Baiti, I.M.; Hotfilder, M.; Lowel, D.; von Luettichau, I.; Mossbrugger, I.; et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 5324–5329.
- Gangwal, K.; Sankar, S.; Hollenhorst, P.C.; Kinsey, M.; Haroldsen, S.C.; Shah, A.A.; Boucher, K.M.; Watkins, W.S.; Jorde, L.B.; Graves, B.J.; et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc. Natl. Acad. Sci. USA 2008, 105, 10149–10154.
- Gangwal, K.; Lessnick, S.L. Microsatellites are EWS/FLI response elements: Genomic “junk” is EWS/FLI’s treasure. Cell Cycle 2008, 7, 3127–3132.
- Guillon, N.; Tirode, F.; Boeva, V.; Zynovyev, A.; Barillot, E.; Delattre, O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS ONE 2009, 4, e4932.
- Yang, L.; Chansky, H.A.; Hickstein, D.D. EWS.Fli-1 fusion protein interacts with hyperphosphorylated RNA polymerase II and interferes with serine-arginine protein-mediated RNA splicing. J. Biol. Chem. 2000, 275, 37612–37618.
- Ahmed, N.S.; Harrell, L.M.; Wieland, D.R.; Lay, M.A.; Thompson, V.F.; Schwartz, J.C. Fusion protein EWS-FLI1 is incorporated into a protein granule in cells. RNA 2021, 27, 920–932.
- Petermann, R.; Mossier, B.M.; Aryee, D.N.; Khazak, V.; Golemis, E.A.; Kovar, H. Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene 1998, 17, 603–610.
- Zhou, H.; Lee, K.A. An hsRPB4/7-dependent yeast assay for trans-activation by the EWS oncogene. Oncogene 2001, 20, 1519–1524.
- Bilke, S.; Schwentner, R.; Yang, F.; Kauer, M.; Jug, G.; Walker, R.L.; Davis, S.; Zhu, Y.J.; Pineda, M.; Meltzer, P.S.; et al. Oncogenic ETS fusions deregulate E2F3 target genes in Ewing sarcoma and prostate cancer. Genome Res. 2013, 23, 1797–1809.
- Schwentner, R.; Papamarkou, T.; Kauer, M.O.; Stathopoulos, V.; Yang, F.; Bilke, S.; Meltzer, P.S.; Girolami, M.; Kovar, H. EWS-FLI1 employs an E2F switch to drive target gene expression. Nucleic Acids Res. 2015, 43, 2780–2789.
- Mertens, F.; Antonescu, C.R.; Mitelman, F. Gene fusions in soft tissue tumors: Recurrent and overlapping pathogenetic themes. Genes Chromosomes Cancer 2016, 55, 291–310.
- Ramakrishnan, R.; Fujimura, Y.; Zou, J.P.; Liu, F.; Lee, L.; Rao, V.N.; Reddy, E.S. Role of protein-protein interactions in the antiapoptotic function of EWS-Fli-1. Oncogene 2004, 23, 7087–7094.
- Harlow, M.L.; Chasse, M.H.; Boguslawski, E.A.; Sorensen, K.M.; Gedminas, J.M.; Kitchen-Goosen, S.M.; Rothbart, S.B.; Taslim, C.; Lessnick, S.L.; Peck, A.S.; et al. Trabectedin Inhibits EWS-FLI1 and Evicts SWI/SNF from Chromatin in a Schedule-dependent Manner. Clin. Cancer Res. 2019, 25, 3417–3429.
- Monument, M.J.; Johnson, K.M.; McIlvaine, E.; Abegglen, L.; Watkins, W.S.; Jorde, L.B.; Womer, R.B.; Beeler, N.; Monovich, L.; Lawlor, E.R.; et al. Clinical and biochemical function of polymorphic NR0B1 GGAA-microsatellites in Ewing sarcoma: A report from the Children’s Oncology Group. PLoS ONE 2014, 9, e104378.
- Lin, L.; Huang, M.; Shi, X.; Mayakonda, A.; Hu, K.; Jiang, Y.Y.; Guo, X.; Chen, L.; Pang, B.; Doan, N.; et al. Super-enhancer-associated MEIS1 promotes transcriptional dysregulation in Ewing sarcoma in co-operation with EWS-FLI1. Nucleic Acids Res. 2019, 47, 1255–1267.
- Sanchez-Molina, S.; Figuerola-Bou, E.; Blanco, E.; Sanchez-Jimenez, M.; Taboas, P.; Gomez, S.; Ballare, C.; Garcia-Dominguez, D.J.; Prada, E.; Hontecillas-Prieto, L.; et al. RING1B recruits EWSR1-FLI1 and cooperates in the remodeling of chromatin necessary for Ewing sarcoma tumorigenesis. Sci. Adv. 2020, 6.
- Smith, R.; Owen, L.A.; Trem, D.J.; Wong, J.S.; Whangbo, J.S.; Golub, T.R.; Lessnick, S.L. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell 2006, 9, 405–416.
- Kinsey, M.; Smith, R.; Lessnick, S.L. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing’s sarcoma. Mol. Cancer Res. 2006, 4, 851–859.
- Boro, A.; Pretre, K.; Rechfeld, F.; Thalhammer, V.; Oesch, S.; Wachtel, M.; Schafer, B.W.; Niggli, F.K. Small-molecule screen identifies modulators of EWS/FLI1 target gene expression and cell survival in Ewing’s sarcoma. Int. J. Cancer 2012, 131, 2153–2164.
- Garcia-Aragoncillo, E.; Carrillo, J.; Lalli, E.; Agra, N.; Gomez-Lopez, G.; Pestana, A.; Alonso, J. DAX1, a direct target of EWS/FLI1 oncoprotein, is a principal regulator of cell-cycle progression in Ewing’s tumor cells. Oncogene 2008, 27, 6034–6043.
- Cironi, L.; Riggi, N.; Provero, P.; Wolf, N.; Suva, M.L.; Suva, D.; Kindler, V.; Stamenkovic, I. IGF1 is a common target gene of Ewing’s sarcoma fusion proteins in mesenchymal progenitor cells. PLoS ONE 2008, 3, e2634.
- Wiles, E.T.; Lui-Sargent, B.; Bell, R.; Lessnick, S.L. BCL11B is up-regulated by EWS/FLI and contributes to the transformed phenotype in Ewing sarcoma. PLoS ONE 2013, 8, e59369.
- Beauchamp, E.; Bulut, G.; Abaan, O.; Chen, K.; Merchant, A.; Matsui, W.; Endo, Y.; Rubin, J.S.; Toretsky, J.; Uren, A. GLI1 is a direct transcriptional target of EWS-FLI1 oncoprotein. J. Biol. Chem. 2009, 284, 9074–9082.
- Abaan, O.D.; Levenson, A.; Khan, O.; Furth, P.A.; Uren, A.; Toretsky, J.A. PTPL1 is a direct transcriptional target of EWS-FLI1 and modulates Ewing’s Sarcoma tumorigenesis. Oncogene 2005, 24, 2715–2722.
- Luo, W.; Xu, C.; Ayello, J.; Dela Cruz, F.; Rosenblum, J.M.; Lessnick, S.L.; Cairo, M.S. Protein phosphatase 1 regulatory subunit 1A in ewing sarcoma tumorigenesis and metastasis. Oncogene 2018, 37, 798–809.
- Grunewald, T.G.; Bernard, V.; Gilardi-Hebenstreit, P.; Raynal, V.; Surdez, D.; Aynaud, M.M.; Mirabeau, O.; Cidre-Aranaz, F.; Tirode, F.; Zaidi, S.; et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat. Genet. 2015, 47, 1073–1078.
- Luo, W.; Gangwal, K.; Sankar, S.; Boucher, K.M.; Thomas, D.; Lessnick, S.L. GSTM4 is a microsatellite-containing EWS/FLI target involved in Ewing’s sarcoma oncogenesis and therapeutic resistance. Oncogene 2009, 28, 4126–4132.
- Charville, G.W.; Wang, W.L.; Ingram, D.R.; Roy, A.; Thomas, D.; Patel, R.M.; Hornick, J.L.; van de Rijn, M.; Lazar, A.J. EWSR1 fusion proteins mediate PAX7 expression in Ewing sarcoma. Mod. Pathol. 2017, 30, 1312–1320.
- Von Heyking, K.; Calzada-Wack, J.; Gollner, S.; Neff, F.; Schmidt, O.; Hensel, T.; Schirmer, D.; Fasan, A.; Esposito, I.; Muller-Tidow, C.; et al. The endochondral bone protein CHM1 sustains an undifferentiated, invasive phenotype, promoting lung metastasis in Ewing sarcoma. Mol. Oncol. 2017, 11, 1288–1301.
- Zhou, Z.; Yu, L.; Kleinerman, E.S. EWS-FLI-1 regulates the neuronal repressor gene REST, which controls Ewing sarcoma growth and vascular morphology. Cancer 2014, 120, 579–588.
- Grunewald, T.G.; Diebold, I.; Esposito, I.; Plehm, S.; Hauer, K.; Thiel, U.; da Silva-Buttkus, P.; Neff, F.; Unland, R.; Muller-Tidow, C.; et al. STEAP1 is associated with the invasive and oxidative stress phenotype of Ewing tumors. Mol. Cancer Res. 2012, 10, 52–65.
- Grunewald, T.G.P.; Ranft, A.; Esposito, I.; da Silva-Buttkus, P.; Aichler, M.; Baumhoer, D.; Schaefer, K.L.; Ottaviano, L.; Poremba, C.; Jundt, G.; et al. High STEAP1 expression is associated with improved outcome of Ewing’s sarcoma patients. Ann. Oncol. 2012, 23, 2185–2190.
- Tang, S.W.; Bilke, S.; Cao, L.; Murai, J.; Sousa, F.G.; Yamade, M.; Rajapakse, V.; Varma, S.; Helman, L.J.; Khan, J.; et al. SLFN11 Is a Transcriptional Target of EWS-FLI1 and a Determinant of Drug Response in Ewing Sarcoma. Clin. Cancer Res. 2015, 21, 4184–4193.
- Ma, Y.; Baltezor, M.; Rajewski, L.; Crow, J.; Samuel, G.; Staggs, V.S.; Chastain, K.M.; Toretsky, J.A.; Weir, S.J.; Godwin, A.K. Targeted inhibition of histone deacetylase leads to suppression of Ewing sarcoma tumor growth through an unappreciated EWS-FLI1/HDAC3/HSP90 signaling axis. J. Mol. Med. 2019, 97, 957–972.
- He, S.; Huang, Q.; Hu, J.; Li, L.; Xiao, Y.; Yu, H.; Han, Z.; Wang, T.; Zhou, W.; Wei, H.; et al. EWS-FLI1-mediated tenascin-C expression promotes tumour progression by targeting MALAT1 through integrin alpha5beta1-mediated YAP activation in Ewing sarcoma. Br. J. Cancer 2019, 121, 922–933.
- Zhang, H.F.; Hughes, C.S.; Li, W.; He, J.Z.; Surdez, D.; El-Naggar, A.M.; Cheng, H.; Prudova, A.; Delaidelli, A.; Negri, G.L.; et al. Proteomic screens for suppressors of anoikis identify IL1RAP as a promising surface target in Ewing sarcoma. Cancer Discov. 2021, 11, 2884–2903.
- Grohar, P.J.; Woldemichael, G.M.; Griffin, L.B.; Mendoza, A.; Chen, Q.R.; Yeung, C.; Currier, D.G.; Davis, S.; Khanna, C.; Khan, J.; et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J. Natl. Cancer Inst. 2011, 103, 962–978.
- Bailly, R.A.; Bosselut, R.; Zucman, J.; Cormier, F.; Delattre, O.; Roussel, M.; Thomas, G.; Ghysdael, J. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol. Cell. Biol. 1994, 14, 3230–3241.
- Li, J.; Ohmura, S.; Marchetto, A.; Orth, M.F.; Imle, R.; Dallmayer, M.; Musa, J.; Knott, M.M.L.; Holting, T.L.B.; Stein, S.; et al. Therapeutic targeting of the PLK1-PRC1-axis triggers cell death in genomically silent childhood cancer. Nat. Commun. 2021, 12, 5356.
- Owen, L.A.; Kowalewski, A.A.; Lessnick, S.L. EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing’s sarcoma. PLoS ONE 2008, 3, e1965.
- Shi, X.; Zheng, Y.; Jiang, L.; Zhou, B.; Yang, W.; Li, L.; Ding, L.; Huang, M.; Gery, S.; Lin, D.C.; et al. EWS-FLI1 regulates and cooperates with core regulatory circuitry in Ewing sarcoma. Nucleic Acids Res. 2020, 48, 11434–11451.
- Markey, F.B.; Romero, B.; Parashar, V.; Batish, M. Identification of a New Transcriptional Co-Regulator of STEAP1 in Ewing’s Sarcoma. Cells 2021, 10, 1300.
- Kinsey, M.; Smith, R.; Iyer, A.K.; McCabe, E.R.; Lessnick, S.L. EWS/FLI and its downstream target NR0B1 interact directly to modulate transcription and oncogenesis in Ewing’s sarcoma. Cancer Res. 2009, 69, 9047–9055.
- Tang, S.W.; Thomas, A.; Murai, J.; Trepel, J.B.; Bates, S.E.; Rajapakse, V.N.; Pommier, Y. Overcoming Resistance to DNA-Targeted Agents by Epigenetic Activation of Schlafen 11 (SLFN11) Expression with Class I Histone Deacetylase Inhibitors. Clin. Cancer Res. 2018, 24, 1944–1953.
- Prieur, A.; Tirode, F.; Cohen, P.; Delattre, O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol. Cell. Biol. 2004, 24, 7275–7283.
- Sankar, S.; Bell, R.; Stephens, B.; Zhuo, R.; Sharma, S.; Bearss, D.J.; Lessnick, S.L. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene 2013, 32, 5089–5100.
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304.
- Lai, A.Y.; Wade, P.A. Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer 2011, 11, 588–596.
- Agra, N.; Cidre, F.; Garcia-Garcia, L.; de la Parra, J.; Alonso, J. Lysyl oxidase is downregulated by the EWS/FLI1 oncoprotein and its propeptide domain displays tumor supressor activities in Ewing sarcoma cells. PLoS ONE 2013, 8, e66281.
- Katschnig, A.M.; Kauer, M.O.; Schwentner, R.; Tomazou, E.M.; Mutz, C.N.; Linder, M.; Sibilia, M.; Alonso, J.; Aryee, D.N.T.; Kovar, H. EWS-FLI1 perturbs MRTFB/YAP-1/TEAD target gene regulation inhibiting cytoskeletal autoregulatory feedback in Ewing sarcoma. Oncogene 2017, 36, 5995–6005.
- Niedan, S.; Kauer, M.; Aryee, D.N.; Kofler, R.; Schwentner, R.; Meier, A.; Potschger, U.; Kontny, U.; Kovar, H. Suppression of FOXO1 is responsible for a growth regulatory repressive transcriptional sub-signature of EWS-FLI1 in Ewing sarcoma. Oncogene 2014, 33, 3927–3938.
- Dylla, L.; Moore, C.; Jedlicka, P. MicroRNAs in Ewing Sarcoma. Front. Oncol. 2013, 3, 65.
- Riggi, N.; Suva, M.L.; De Vito, C.; Provero, P.; Stehle, J.C.; Baumer, K.; Cironi, L.; Janiszewska, M.; Petricevic, T.; Suva, D.; et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010, 24, 916–932.
- Robin, T.P.; Smith, A.; McKinsey, E.; Reaves, L.; Jedlicka, P.; Ford, H.L. EWS/FLI1 regulates EYA3 in Ewing sarcoma via modulation of miRNA-708, resulting in increased cell survival and chemoresistance. Mol. Cancer Res. 2012, 10, 1098–1108.
- Sparmann, A.; van Lohuizen, M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 2006, 6, 846–856.
- Nakatani, F.; Ferracin, M.; Manara, M.C.; Ventura, S.; Del Monaco, V.; Ferrari, S.; Alberghini, M.; Grilli, A.; Knuutila, S.; Schaefer, K.L.; et al. miR-34a predicts survival of Ewing’s sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J. Pathol. 2012, 226, 796–805.
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191.
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016.
- Wang, J.; Lee, J.E.; Riemondy, K.; Yu, Y.; Marquez, S.M.; Lai, E.C.; Yi, R. XPO5 promotes primary miRNA processing independently of RanGTP. Nat. Commun. 2020, 11, 1845.
- Selvanathan, S.P.; Graham, G.T.; Erkizan, H.V.; Dirksen, U.; Natarajan, T.G.; Dakic, A.; Yu, S.; Liu, X.; Paulsen, M.T.; Ljungman, M.E.; et al. Oncogenic fusion protein EWS-FLI1 is a network hub that regulates alternative splicing. Proc. Natl. Acad. Sci. USA 2015, 112, E1307–E1316.
- Sun, H.L.; Cui, R.; Zhou, J.; Teng, K.Y.; Hsiao, Y.H.; Nakanishi, K.; Fassan, M.; Luo, Z.; Shi, G.; Tili, E.; et al. ERK Activation Globally Downregulates miRNAs through Phosphorylating Exportin-5. Cancer Cell 2016, 30, 723–736.
- Klevernic, I.V.; Morton, S.; Davis, R.J.; Cohen, P. Phosphorylation of Ewing’s sarcoma protein (EWS) and EWS-Fli1 in response to DNA damage. Biochem. J. 2009, 418, 625–634.
- Bachmaier, R.; Aryee, D.N.; Jug, G.; Kauer, M.; Kreppel, M.; Lee, K.A.; Kovar, H. O-GlcNAcylation is involved in the transcriptional activity of EWS-FLI1 in Ewing’s sarcoma. Oncogene 2009, 28, 1280–1284.
- Olsen, R.J.; Hinrichs, S.H. Phosphorylation of the EWS IQ domain regulates transcriptional activity of the EWS/ATF1 and EWS/FLI1 fusion proteins. Oncogene 2001, 20, 1756–1764.
- Schlottmann, S.; Erkizan, H.V.; Barber-Rotenberg, J.S.; Knights, C.; Cheema, A.; Uren, A.; Avantaggiati, M.L.; Toretsky, J.A. Acetylation Increases EWS-FLI1 DNA Binding and Transcriptional Activity. Front. Oncol. 2012, 2, 107.
- Maniatis, T.; Tasic, B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 2002, 418, 236–243.
- Eymin, B. Targeting the spliceosome machinery: A new therapeutic axis in cancer? Biochem. Pharmacol. 2021, 189, 114039.
- Embree, L.J.; Azuma, M.; Hickstein, D.D. Ewing sarcoma fusion protein EWSR1/FLI1 interacts with EWSR1 leading to mitotic defects in zebrafish embryos and human cell lines. Cancer Res. 2009, 69, 4363–4371.
- Knoop, L.L.; Baker, S.J. The splicing factor U1C represses EWS/FLI-mediated transactivation. J. Biol. Chem. 2000, 275, 24865–24871.
- Selvanathan, S.P.; Graham, G.T.; Grego, A.R.; Baker, T.M.; Hogg, J.R.; Simpson, M.; Batish, M.; Crompton, B.; Stegmaier, K.; Tomazou, E.M.; et al. EWS-FLI1 modulated alternative splicing of ARID1A reveals novel oncogenic function through the BAF complex. Nucleic Acids Res. 2019, 47, 9619–9636.
- Hensel, T.; Giorgi, C.; Schmidt, O.; Calzada-Wack, J.; Neff, F.; Buch, T.; Niggli, F.K.; Schafer, B.W.; Burdach, S.; Richter, G.H. Targeting the EWS-ETS transcriptional program by BET bromodomain inhibition in Ewing sarcoma. Oncotarget 2016, 7, 1451–1463.
- García-Domínguez, D.J.; Hajji, N.; Sánchez-Molina, S.; Figuerola-Bou, E.; de Pablos, R.M.; Espinosa-Oliva, A.M.; Andrés-León, E.; Terrón-Camero, L.C.; Flores-Campos, R.; Pascual-Pasto, G.; et al. HDAC6 regulates expression of the oncogenic driver EWSR1-FLI1 through the <em>EWSR1</em> promoter in Ewing sarcoma. bioRxiv 2021.
- Ban, J.; Jug, G.; Mestdagh, P.; Schwentner, R.; Kauer, M.; Aryee, D.N.; Schaefer, K.L.; Nakatani, F.; Scotlandi, K.; Reiter, M.; et al. Hsa-mir-145 is the top EWS-FLI1-repressed microRNA involved in a positive feedback loop in Ewing’s sarcoma. Oncogene 2011, 30, 2173–2180.
- Keskin, T.; Bakaric, A.; Waszyk, P.; Boulay, G.; Torsello, M.; Cornaz-Buros, S.; Chevalier, N.; Geiser, T.; Martin, P.; Volorio, A.; et al. LIN28B Underlies the Pathogenesis of a Subclass of Ewing Sarcoma LIN28B Control of EWS-FLI1 Stability. Cell Rep. 2020, 30, 4567–4583.e5.
- De Vito, C.; Riggi, N.; Suva, M.L.; Janiszewska, M.; Horlbeck, J.; Baumer, K.; Provero, P.; Stamenkovic, I. Let-7a is a direct EWS-FLI-1 target implicated in Ewing’s sarcoma development. PLoS ONE 2011, 6, e23592.
- Gierisch, M.E.; Pfistner, F.; Lopez-Garcia, L.A.; Harder, L.; Schafer, B.W.; Niggli, F.K. Proteasomal Degradation of the EWS-FLI1 Fusion Protein Is Regulated by a Single Lysine Residue. J. Biol. Chem. 2016, 291, 26922–26933.
- Gierisch, M.E.; Pedot, G.; Walser, F.; Lopez-Garcia, L.A.; Jaaks, P.; Niggli, F.K.; Schafer, B.W. USP19 deubiquitinates EWS-FLI1 to regulate Ewing sarcoma growth. Sci. Rep. 2019, 9, 951.
- Seong, B.K.A.; Dharia, N.V.; Lin, S.; Donovan, K.A.; Chong, S.; Robichaud, A.; Conway, A.; Hamze, A.; Ross, L.; Alexe, G.; et al. TRIM8 modulates the EWS/FLI oncoprotein to promote survival in Ewing sarcoma. Cancer Cell 2021, 39, 1262–1278.e1267.
- Su, S.; Chen, J.; Jiang, Y.; Wang, Y.; Vital, T.; Zhang, J.; Laggner, C.; Nguyen, K.T.; Zhu, Z.; Prevatte, A.W.; et al. SPOP and OTUD7A Control EWS-FLI1 Protein Stability to Govern Ewing Sarcoma Growth. Adv. Sci. 2021, 8, e2004846.
- Sun, H.; Lin, D.C.; Cao, Q.; Guo, X.; Marijon, H.; Zhao, Z.; Gery, S.; Xu, L.; Yang, H.; Pang, B.; et al. CRM1 Inhibition Promotes Cytotoxicity in Ewing Sarcoma Cells by Repressing EWS-FLI1-Dependent IGF-1 Signaling. Cancer Res. 2016, 76, 2687–2697.
- Bailey, K.M.; Julian, C.M.; Klinghoffer, A.N.; Bernard, H.; Lucas, P.C.; McAllister-Lucas, L.M. EWS-FLI1 low Ewing sarcoma cells demonstrate decreased susceptibility to T-cell-mediated tumor cell apoptosis. Oncotarget 2019, 10, 3385–3399.
- Sengupta, A.; Rahman, M.; Mateo-Lozano, S.; Tirado, O.M.; Notario, V. The dual inhibitory effect of thiostrepton on FoxM1 and EWS/FLI1 provides a novel therapeutic option for Ewing’s sarcoma. Int. J. Oncol. 2013, 43, 803–812.
- Stegmaier, K.; Wong, J.S.; Ross, K.N.; Chow, K.T.; Peck, D.; Wright, R.D.; Lessnick, S.L.; Kung, A.L.; Golub, T.R. Signature-based small molecule screening identifies cytosine arabinoside as an EWS/FLI modulator in Ewing sarcoma. PLoS Med. 2007, 4, e122.
- DuBois, S.G.; Krailo, M.D.; Lessnick, S.L.; Smith, R.; Chen, Z.; Marina, N.; Grier, H.E.; Stegmaier, K.; Children’s Oncology, G. Phase II study of intermediate-dose cytarabine in patients with relapsed or refractory Ewing sarcoma: A report from the Children’s Oncology Group. Pediatr. Blood Cancer 2009, 52, 324–327.
- Chu, Z.; Gu, L.; Hu, Y.; Zhang, X.; Li, M.; Chen, J.; Teng, D.; Huang, M.; Shen, C.H.; Cai, L.; et al. STAG2 regulates interferon signaling in melanoma via enhancer loop reprogramming. Nat. Commun. 2022, 13, 1859.
- Tirode, F.; Surdez, D.; Ma, X.; Parker, M.; Le Deley, M.C.; Bahrami, A.; Zhang, Z.; Lapouble, E.; Grossetete-Lalami, S.; Rusch, M.; et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014, 4, 1342–1353.
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018, 174, 1034–1035.
- Adane, B.; Alexe, G.; Seong, B.K.A.; Lu, D.; Hwang, E.E.; Hnisz, D.; Lareau, C.A.; Ross, L.; Lin, S.; Dela Cruz, F.S.; et al. STAG2 loss rewires oncogenic and developmental programs to promote metastasis in Ewing sarcoma. Cancer Cell 2021, 39, 827–844.e810.
- Surdez, D.; Zaidi, S.; Grossetete, S.; Laud-Duval, K.; Ferre, A.S.; Mous, L.; Vourc’h, T.; Tirode, F.; Pierron, G.; Raynal, V.; et al. STAG2 mutations alter CTCF-anchored loop extrusion, reduce cis-regulatory interactions and EWSR1-FLI1 activity in Ewing sarcoma. Cancer Cell 2021, 39, 810–826.e819.
- Crompton, B.D.; Stewart, C.; Taylor-Weiner, A.; Alexe, G.; Kurek, K.C.; Calicchio, M.L.; Kiezun, A.; Carter, S.L.; Shukla, S.A.; Mehta, S.S.; et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014, 4, 1326–1341.
- Lu, D.Y.; Ellegast, J.M.; Ross, K.N.; Malone, C.F.; Lin, S.; Mabe, N.W.; Dharia, N.V.; Meyer, A.; Conway, A.; Su, A.H.; et al. The ETS transcription factor ETV6 constrains the transcriptional activity of EWS-FLI to promote Ewing sarcoma. Nat. Cell Biol. 2023, 25, 285–297.
- Gao, Y.; He, X.Y.; Wu, X.S.; Huang, Y.H.; Toneyan, S.; Ha, T.; Ipsaro, J.J.; Koo, P.K.; Joshua-Tor, L.; Bailey, K.M.; et al. ETV6 dependency in Ewing sarcoma by antagonism of EWS-FLI1-mediated enhancer activation. Nat. Cell Biol. 2023, 25, 298–308.
- Berghuis, D.; de Hooge, A.S.; Santos, S.J.; Horst, D.; Wiertz, E.J.; van Eggermond, M.C.; van den Elsen, P.J.; Taminiau, A.H.; Ottaviano, L.; Schaefer, K.L.; et al. Reduced human leukocyte antigen expression in advanced-stage Ewing sarcoma: Implications for immune recognition. J. Pathol. 2009, 218, 222–231.
- Morales, E.; Olson, M.; Iglesias, F.; Dahiya, S.; Luetkens, T.; Atanackovic, D. Role of immunotherapy in Ewing sarcoma. J. Immunother. Cancer 2020, 8, e000653.
- Englisch, A.; Altvater, B.; Kailayangiri, S.; Hartmann, W.; Rossig, C. VEGFR2 as a target for CAR T cell therapy of Ewing sarcoma. Pediatr. Blood Cancer 2020, 67, e28313.
- Challita-Eid, P.M.; Morrison, K.; Etessami, S.; An, Z.; Morrison, K.J.; Perez-Villar, J.J.; Raitano, A.B.; Jia, X.C.; Gudas, J.M.; Kanner, S.B.; et al. Monoclonal antibodies to six-transmembrane epithelial antigen of the prostate-1 inhibit intercellular communication in vitro and growth of human tumor xenografts in vivo. Cancer Res. 2007, 67, 5798–5805.
- Richter, G.H.; Fasan, A.; Hauer, K.; Grunewald, T.G.; Berns, C.; Rossler, S.; Naumann, I.; Staege, M.S.; Fulda, S.; Esposito, I.; et al. G-Protein coupled receptor 64 promotes invasiveness and metastasis in Ewing sarcomas through PGF and MMP1. J. Pathol. 2013, 230, 70–81.
- Huang, X.; Park, H.; Greene, J.; Pao, J.; Mulvey, E.; Zhou, S.X.; Albert, C.M.; Moy, F.; Sachdev, D.; Yee, D.; et al. IGF1R- and ROR1-Specific CAR T Cells as a Potential Therapy for High Risk Sarcomas. PLoS ONE 2015, 10, e0133152.
- Schirmer, D.; Storz, I.; Wisskirchen, K.; Feederle, R.; Schmidt, O.; Abken, H.; Protzer, U.; Burdach, S.; Richter, G.H.S. GPR64-specific CAR-transgenic T cells selectively kill Ewing sarcoma in vivo. Oncol. Res. Treat. 2018, 41, 130–131.
- Town, J.; Pais, H.; Harrison, S.; Stead, L.F.; Bataille, C.; Bunjobpol, W.; Zhang, J.; Rabbitts, T.H. Exploring the surfaceome of Ewing sarcoma identifies a new and unique therapeutic target. Proc. Natl. Acad. Sci. USA 2016, 113, 3603–3608.
- Kailayangiri, S.; Altvater, B.; Lesch, S.; Balbach, S.; Gottlich, C.; Kuhnemundt, J.; Mikesch, J.H.; Schelhaas, S.; Jamitzky, S.; Meltzer, J.; et al. EZH2 Inhibition in Ewing Sarcoma Upregulates G(D2) Expression for Targeting with Gene-Modified T Cells. Mol. Ther. 2019, 27, 933–946.
More
Information
Subjects:
Oncology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
2 times
(View History)
Update Date:
17 Aug 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




