Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Gianluigi Giannelli | -- | 4736 | 2023-08-16 17:43:05 | | | |
| 2 | Lindsay Dong | Meta information modification | 4736 | 2023-08-17 03:19:54 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Giannelli, G.; Potestio, S.; Visioli, G. Plant-Growth-Promoting Rhizobacteria in Salt Stress Tolerance in Crops. Encyclopedia. Available online: https://encyclopedia.pub/entry/48137 (accessed on 08 February 2026).
Giannelli G, Potestio S, Visioli G. Plant-Growth-Promoting Rhizobacteria in Salt Stress Tolerance in Crops. Encyclopedia. Available at: https://encyclopedia.pub/entry/48137. Accessed February 08, 2026.
Giannelli, Gianluigi, Silvia Potestio, Giovanna Visioli. "Plant-Growth-Promoting Rhizobacteria in Salt Stress Tolerance in Crops" Encyclopedia, https://encyclopedia.pub/entry/48137 (accessed February 08, 2026).
Giannelli, G., Potestio, S., & Visioli, G. (2023, August 16). Plant-Growth-Promoting Rhizobacteria in Salt Stress Tolerance in Crops. In Encyclopedia. https://encyclopedia.pub/entry/48137
Giannelli, Gianluigi, et al. "Plant-Growth-Promoting Rhizobacteria in Salt Stress Tolerance in Crops." Encyclopedia. Web. 16 August, 2023.
Copy Citation
The use of beneficial microorganisms inhabiting the rhizosphere (PGPR) is a promising tool to alleviate salt stress in various crops and represents a strategy to increase agricultural productivity in salt soils. Increasing evidence underlines that PGPR affect plant physiological, biochemical, and molecular responses to salt stress. The mechanisms behind these phenomena include osmotic adjustment, modulation of the plant antioxidant system, ion homeostasis, modulation of the phytohormonal balance, increase in nutrient uptake, and the formation of biofilms
plant-growth-promoting rhizobacteria (PGPR)
salt stress
plant salt-responsive genes
PGPR plant gene modulation
1. Rhizosphere Microbial Community
Since their earliest evolution, land plants have been associated with a complex microbial community that includes beneficial microorganisms. The presence of beneficial microorganisms helped early land plants to respond to the environment and face challenges such as access to nutrients, new and/or stressful conditions, and pathogens [1][2].
In soil, there is a gradient of intimacy between plant roots and microorganisms, and the plant’s influence on the microbial community is greater near the root surface. A volume of soil specially influenced by plant roots or associated with the roots and plant-produced material is defined as a rhizosphere [3], a term originally coined by Hiltner [4] to describe the microorganisms of soil around and inside roots. Microorganisms living on the surface of roots are now said to inhabit the rhizosphere and the rhizoplane, and those living inside the roots are called endophytes [5][6].
Among the microbial communities colonizing the different plant regions (i.e., flowers, fruits, stems, leaves, and roots), the one associated with the roots (rhizobiome) is undoubtedly the most populated and most sophisticated of all microbial communities associated with high plants [1]. The rhizosphere effect is caused by the enormous influence exerted by the plant root exudates, a phenomenon called rhizodeposition, including a range of organic acids, amino acids, sugars, nucleosides, vitamins, and other small molecules that act as strong chemo-attractants of the soil microbiota [7]. Rhizodeposition also refers to the release into the rhizosphere of a group of specialized cells, called root cap boundary cells. This cell type is considered crucial for its effect on the rhizosphere as it usually survives after it has decayed from the roots [8][9] In particular, the passive flux of low-molecular-weight metabolites such as sugars, amino acids, and organic acids through root exudation is mostly located at the root tip, where the lack of cell differentiation favors the diffusion of metabolites to the soil [10].
The root tip senses changes in the concentration of exuded metabolites and translates them into signals to modify root growth. Through root exudate flux, plants can locally enhance concentrations of many common metabolites, which can serve as sensors and integrators of the plant nutritional status and of the nutrient availability for the rhizosphere microbial community [11][12]. Plant-associated microorganisms also constitute a strong sink for plant carbon, thereby increasing concentration gradients of metabolites and affecting root exudation. Indeed, it was found that when microorganisms were present in plant growth solution, exudation was enhanced compared to axenic conditions [13]. Rhizodeposits account for approximately 11% of the net carbon fixed by photosynthesis, and 10–16% of the total nitrogen in plants, but these values vary greatly depending on plant species and age [14]. The net sequestration of organic carbon and nitrogen by roots stimulates the multiplication of soil microbes near root tissues because (i) most known soil bacteria are organotrophic (i.e., they obtain growth energy from organic substrates) and (ii) in most soils, the access and availability of organic compounds are limited.
The microbial population colonizing the rhizosphere includes bacteria, which are the most abundant, as well as fungi, protozoa, and algae, and its composition is markedly different from that of its surrounding soil [15][16]. Regarding bacteria, Weller and Thomashow [17] proved that their presence in the rhizosphere is generally 10 to 100 times greater than in bulk soil. All plant–microbe interactions can have mutualistic or detrimental effects on the host during the microbes’ attempts to obtain nutrients and environmental protection, regardless of the physical interaction [18]. For this reason, plants need a selection mechanism that favors rhizosphere colonization by beneficial and non-pathogenic microorganisms.
The literature available suggests a two-step selection process that gradually distinguishes the root microbiota from the surrounding soil biomes and determines differences in the dimension and composition of the microbial community. In the first step, it is suggested that rhizodeposition and the recognition of the host cell wall promote organotrophic bacteria growth, thereby initiating the first community shift of the soil biome. In the second step, the microbial subpopulation undergoes a second selection process near and within the roots, based on host genotype-dependent selection, defining the profiles of the microbial communities that prosper in the rhizoplane and within the roots [8][19].
2. Plant-Growth-Promoting Rhizobacteria (PGPR)
In 1978, Kloepper and Schroth [20] introduced the term “rhizobacteria” to indicate the soil bacterial community capable of competitively colonizing plant roots and stimulating growth, thereby reducing the incidence of plant disease. They were then renamed plant-growth-promoting rhizobacteria in 1981 [21]. PGPR represent about 2–5% of the total rhizosphere bacteria [22]. According to Nakkeeran et al. [23], an ideal PGPR should possess high rhizosphere competence, improve plant growth capabilities, have a wide range of action, be safe in relation to the environment, compatible with other rhizobacteria, and tolerant to heat, UV radiation, and oxidants.
Based on the interaction with plants and their degree of association, PGPR can be classified as either intracellular PGPR (iPGPR or symbiotic bacteria), which live inside the plants and exchange metabolites with them directly, or free-living rhizobacteria or extracellular PGPR (ePGPR), which live outside plant cells [5][24]. iPGPR include a wide range of soil bacterial genera that are usually located inside the specialized nodular structures of root cells such as Allorhizobium, Azorhizobium, Bradyrhizobium, Mesorhizobium, and Rhizobium of the family Rhizobiaceae [25]. ePGPR, instead, are present in the rhizosphere, in the rhizoplane, or in the spaces between the root cortex cells, and include genera such as Agrobacterium, Arthrobacter, Azotobacter, Azospirillum, Bacillus, Burkholderia, Caulobacter, Chromobacterium, Erwinia, Flavobacterium, Micrococcous, Pseudomonas, and Serratia [5]. The beneficial action of PGPR is exerted through a series of processes that result in three macro effects on the host plant: (i) increased nutrient assimilation; (ii) increased resistance to phytopathogen infections (viruses, fungi, bacteria, nematodes, insects, etc.), also referred to as biotic stresses; and (iii) increased tolerance to abiotic stresses (heavy metal in soils, drought, nutrient deficiency, salinity, temperature). This leads to improved plant performance and, consequently increased growth. The diverse mechanisms underlying these phenomena include (i) N2 supply through biological nitrogen fixation; (ii) the production of siderophores; (iii) the solubilization and mineralization of nutrients; (iv) the synthesis of ACC (1-aminocyclopropane-1-carboxylate) deaminase; (v) the production and modulation of phytohormones (e.g., indole 3-acetic acid (IAA), cytokinins (CK), abscisic acid (ABA), gibberellic acid (GA)); (vi) the control of plant pathogens through various mechanisms such as the secretion of enzymes able to hydrolase the fungal cell wall or compete for nutrients, the induction of systemic resistance (ISR), and the production of siderophores and antibiotics; (vii) the bio-remediation of heavy-metal-contaminated soils; and viii) an improvement in abiotic stress resistance [26][27][28][29][30].
3. Salt Stress Affects Plant Growth
3.1. Soil Salinization
Soil salinity is a major abiotic stress in agricultural crop productivity worldwide. The causes of soil salinization can be grouped into primary, due to natural sources, and secondary, which are largely anthropogenic and exacerbate the problem of salinization [31][32].
A primary source is saline parent material that releases soil mineral constituents during the chemical erosion of rock or sediment minerals, which react with air and water to produce soluble salts that are then transported away from their source of origin through watercourses. Another factor determining the development of saline soils is salty groundwater that rises through the soil profile via capillarity and releases salts, which then remain in the soil due to water evaporation. Coastal areas are particularly prone to salinization, since winds and sea spray along the coast transport salts from the sea inland in sufficient quantities to cause salinization of these areas. Sea spray can have an impact up to 80 km inland or even beyond [31].
The main secondary cause is irrigation. Irrigation can cause salinity when salty water is used or when it causes the inadequate leaching of salts into the soil. Irrigation water can also recharge underground wells and cause them to rise, gradually introducing the salts contained in groundwater into the soil. The use of fertilized water can also contribute to the introduction of salts, which accumulate over time and increase with repeated application. Waste and wastewater are also secondary contributors to soil salinity, and so is variation in land use or land cover.
Soil is generally considered saline when the electrical conductivity (EC) of the saturation extract in the root zone exceeds 40 mM at 25 °C, with 15% of unbound Na+ ions [33]. Among the different ions that can cause salinization, sodium is the most soluble and widespread, and it is considered the most deleterious. In fact, unlike other ions, such as Ca2+, Mg2+, or K+, which plants can exploit for normal physiological and biochemical processes, Na+ is useless and extremely toxic to glycophytes [34].
Soils provide ecosystem services that are essential to human life and biodiversity. The salinization of soils has a serious impact on these services, as it leads to a number of consequences including (i) a reduction in agricultural productivity, water quality, soil biodiversity, and an increase in soil erosion; (ii) a decreased ability to act against contaminants as buffers and filters; (iii) a degraded soil structure; (iv) decreased functions of ecological systems such as the hydrological and nutrient cycles; (v) an increased concentration of ions that are toxic to plants; (vi) a reduced ability of crops to take up water; and (vii) reduced soil fertility and availability of micronutrients [35].
3.2. Plant Response to Salt Stress
Salinity stress involves changes in various plant physiological and metabolic processes, depending on the severity and duration of the stress, and ultimately inhibits plant growth. High salinity affects plants in several ways: water stress, ion toxicity, nutritional disorders, oxidative stress, the alteration of metabolic processes, membrane disorganization, a reduction in cell division and expansion, and genotoxicity [36].
It is not surprising that plants have evolved mechanisms to regulate NaCl accumulation, since NaCl is the most soluble and widespread salt. Analyses of the different growth responses under salinity conditions reveal that plants differ considerably in their tolerance [37]. Barley is the most tolerant crop among cereals, while rice is the most sensitive. Similarly, soft wheat (Triticum aestivum) is moderately tolerant, whereas durum wheat (Triticum turgidum ssp. Durum) is more sensitive to salt stress. Tall grass (Thinopyrum ponticum, syn. Agropyron elongatum) is a halophytic relative of wheat and is one of the most tolerant plants among monocotyledons.
An elevated salt concentration reduces shoot growth and affects plants in two main ways: it reduces the roots’ osmotic potential, thereby complicating water uptake (osmotic effect), and inside plants, it results in ion toxicity (ionic effect) [38]. A two-phase model has been proposed to describe the effects of salt stress. The osmotic phase begins immediately after the exposure of the roots to a threshold level of salinity, which is usually 40 mM NaCl or less for plants such as Arabidopsis. During this phase, the immediate response is stomatal closure, which also helps to reduce ion flux to the shoot; the rate of shoot growth declines, and new leaves emerge more slowly [36][37]. However, this is an unsustainable solution and is bound to be discontinued in a short time because of the difference in water potential between the atmosphere and the leaf cells, but especially due to the need to fix carbon.
Usually, osmotic stress has an immediate and greater effect on growth rate than ionic stress, except for cases with a high salinity concentration or in sensitive species that do not have a strong ability to control Na+ transport. For most plant species, Na+ toxicity is greater than Cl− toxicity; for this reason, almost all studies have focused on the mechanisms involved in Na+ control rather than Cl− [37].
The main physiological, molecular, and biochemical mechanisms for survival under high salt concentration include (i) ion homeostasis and compartmentalization, (ii) ion transport and uptake, (iii) the biosynthesis of osmoprotectants and compatible solutes, (iv) the activation of the antioxidant enzyme and synthesis of antioxidant compounds, and (v) the modulation of hormones [39][40].
3.3. PGPR Growth and Activity in Salt Stress Conditions
Most of the studies aiming to dissect PGPR activities under salt conditions are conducted in vitro, yet these observations are not directly correlated with soil in nature. Increasing NaCl concentration from 0 to 1.2 M NaCl progressively reduces the amount of ACC deaminase activity, IAA, P-solubilization, and EPS in Bacillus pumilus JPV11 [41]. A 38% reduction in IAA production was observed in B. megaterium when subjected to 5% salinity, while it increases in P. fluorescens cultures. When growing in 2% NaCl, V. paradoxus reduces the production of siderophores. No changes were observed in siderophore production for the other two strains, while a slight reduction was observed when increasing the salt concentration. There was no significant change in the ACC-deaminase activity of P. fluorescens and B. megaterium with salt concentrations, but there was a reduction in V. paradoxus [42].
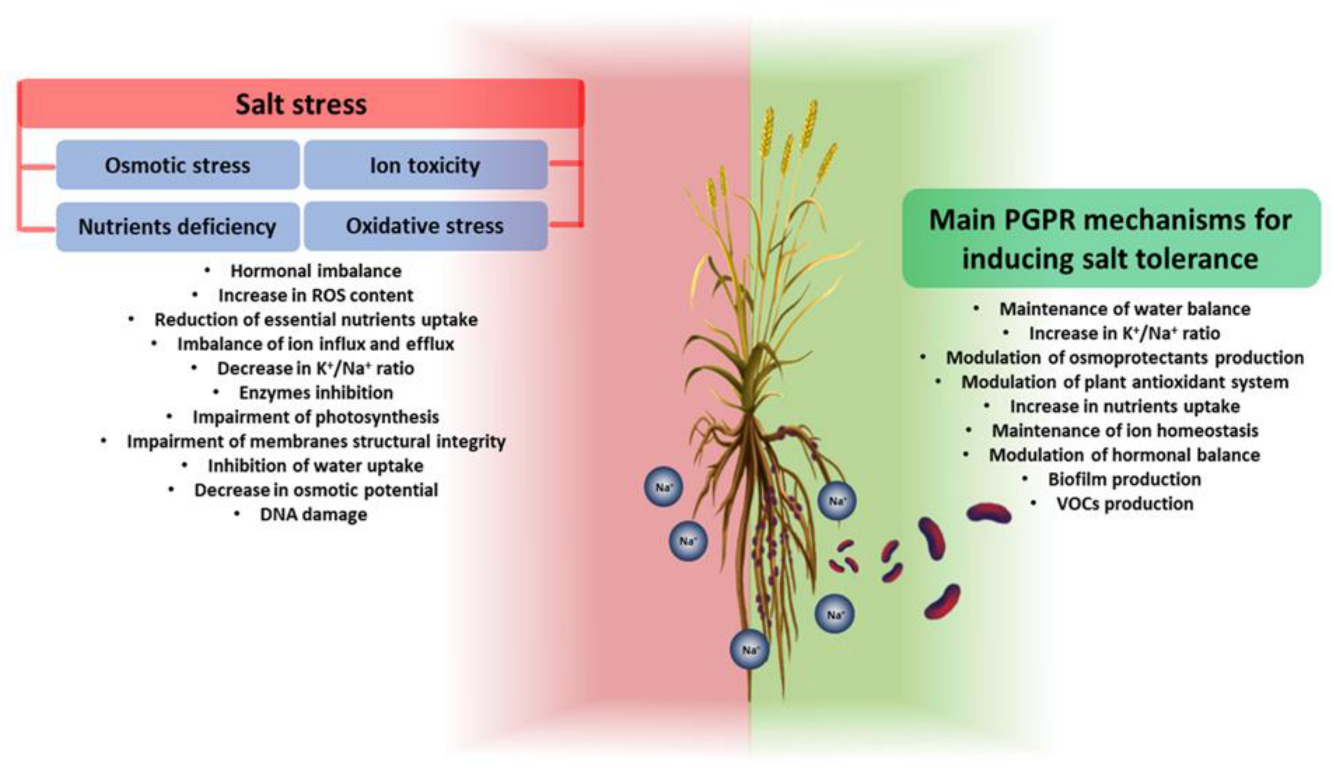
3.4. Contribution of PGPR to Plant Salt Tolerance
PGPR exhibit beneficial traits that allow them to mitigate the toxic effects caused by high salt concentrations. PGPR can improve plant growth in a salt environment in two main ways: (i) activating or modulating the response systems of plants during exposure to salt; and (ii) synthesizing anti-stress molecules [43]. Mechanisms to improve the growth and resistance of plants exposed to salinity include (i) the improvement of nutrient uptake (e.g., N2 fixation, release of bound P and K+ from the soil, chelating iron) and maintenance of the water balance; (ii) an influence on ion homeostasis; (iii) the induction of the selective absorption of K+ and exclusion of Na+ to maintain a high K+/Na+ ratio; (iv) the formation of biofilm to reduce Na+ toxicity; (v) changes in root architecture; (vi) the modulation of the antioxidant system; (vii) the modulation of osmotic substances; (viii) the modulation of plant hormonal levels; and (ix) the modulation of the expression of salt-responsive genes [44]. GPR are classified according to their mechanisms, but in the context of salt stress the analysis of their influence on the plant’s response has shown that their promoting activity is never due to a single mechanism. The roles of PGPR in enhancing plant salt tolerance by modulating different genes and plant cellular functions can be seen in Figure 1.

Figure 1. General overview of salt stress effects on plants (on the left) and PGPR mechanisms involved in plant salt-stress tolerance (on the right). Salt stress negatively affects all plant growth and development processes. Through different mechanisms, PGPR can increase plant tolerance to salt stress.
3.4.1. Osmotic Adjustment
A decrease in osmotic pressure induces water loss and represents one of the problems plants face when growing in saline soils, because changes in osmotic pressure must be compensated for to maintain cell volume and turgor [45]. In order to maintain osmotic balance both inside and outside the cell, plants increase the synthesis of low-molecular-weight compatible solutes in the cytosol, also referred to as osmolytes. The most common osmolytes include proline, glycine betaine, soluble sugars, and ectoine [46][47]. Osmolytes are not charged, polar, and soluble, and do not interfere with cell metabolism even at high concentrations [39]. Osmotic adjustment is an energy-intensive process; in fact, it results in a reduction in growth to direct energy to osmolyte synthesis. On the other hand, it is a necessary measure to alleviate the effects of salt stress [48][49]. Proline also acts as a scavenger for stress-induced ROS; buffers the cell redox potential; and stabilizes proteins, enzymes, membrane structures, and the electron transport system complex II [50][51][52]. Moreover, proline is considered a sensitive physiological marker of salt stress.
In plants, ABA signaling and MAP kinases are involved in the production and accumulation of osmolytes [53]. The rapid activation of MAPKs such as MAPK3, 4, 6 was frequently observed in response to salt stress [54]. For instance, the salt-induced activation of MKK4 regulates the activity of MPK3, which increases the expression of stress-responsive genes such as NCED3 and RD29A [55]. The protein kinase SnRK2 family can be activated in both an ABA-dependent and -independent way. SnRK2.2, SnRK2.3, and SnRK2.6 are important for transducing the ABA signal, activating AREB/ABF TFs (ABA-responsive element binding factors), and regulating the downstream responsive gene [56][57][58].
3.4.2. Antioxidant Activity
Many adverse environmental factors can alter the equilibrium between ROS production and scavenging activity [59]. The content of reactive oxygen species (ROS) increases dramatically during salt stress, mainly due to the disruption of electron transport chains (ETC) during photoinhibition and/or a decrease in water potential [60]. ROS comprise different compounds such as O2•−, H2O2, 1O2, HO2•−, OH•, ROOH, ROO•, and RO•, which react spontaneously with organic molecules and cause membrane lipid peroxidation, protein oxidation, enzyme inhibition, and DNA and RNA damage [61]. In order to detoxify ROS, plants have developed antioxidant systems including antioxidant enzymes and non-enzymatic compounds. Antioxidant enzymes, such as superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPX), ascorbate peroxidase (APX), and glutathione reductase (GR), and the accumulation of non-enzymatic antioxidant compounds (carotenoids, flavonoids and other phenolics, proline) are positively correlated with a tolerance to salinity in plants [39].
MAPK cascades act as important signaling pathways in responding to oxidative stress and regulating ROS homeostasis. For example, the activity of scavenging enzymes is controlled by the MEKK1-MKK1/MKK2-MPK4 cascade to maintain ROS homeostasis. In Arabidopsis, two other proteins, MPK3 and MPK6, phosphorylate and activate HEAT SHOCK FACTOR A4A (HSFA4A), controlling ROS homeostasis and positively regulating the salt stress response [62]. Increasing evidence suggests an active role for PGPR in modulating the antioxidant defense system of plants by increasing the activity of various antioxidant enzymes under stress conditions [63][64][65].
3.4.3. Ion Homeostasis
Regardless of their nature, neither glycophytes nor halophytes can tolerate high concentrations of Na+ in their cytoplasm. The maintenance of ion homeostasis through Na+ ion compartmentalization is critical for growth during salt stress [36]. The regulation of Na+ uptake and transport in salt-stressed plants has been interpreted in the context of maintaining high K+/Na+ ratios in the cytoplasm. Because Na+ and K+ possess very similar physical and chemical properties, the two ions compete for many key metabolic processes in the cytoplasm. In fact, Na+ inhibits the enzymatic activity of many enzymes that require K+ to function. Since more than 50 different cytoplasmic enzymes are activated by K+, the disruption of their activity leads to a dramatic effect on cellular metabolism [66]. Na+ control mechanisms involve Na+ exclusion from roots, Na+ long-distance transport, and Na+ compartmentation [67]. Three main proton pumps have been identified as being related to salt stress tolerance in plants: (i) vacuolar-proton phosphatase which generates a proton gradient by using energy from pyrophosphate (Ppi); (ii) plasma membrane H+/ATPase; and (iii) vacuolar H+/ATPase which couples H+ transport and ATP hydrolysis [68]. The salt overly sensitive (SOS) stress-signaling pathway is considered the first mechanism used by plants to exclude Na+, and increasing evidence demonstrates its role in ion homeostasis and salt tolerance. The SOS signaling pathway is composed of three proteins: SOS3 (Ca2+-binding protein), SOS2 (serine/threonine kinase), and SOS1 (Na+/H+ antiporter). High salinity triggers an increase in cytosolic Ca2+ concentration, inducing the Ca2+-mediated activation of SOS3. The interaction between SOS2 and the SOS3 protein results in the activation of kinase and the release of the SOS3/SOS2 complex into the cytosol, where it phosphorylates SOS1, activating its transport function [38][69]. SOS1 is required to extract excess Na+ from cells (i.e., into the rhizosphere through root epidermal cells, or into the xylem through parenchyma cells of the xylem), and thus reduce ionic stress [70][71].
The Bacillus megaterium ZS-3 endophytic strain induces systemic tolerance to salt stress in Arabidopsis thaliana through the regulation of different processes, including photosynthesis, osmotic adjustment, and ion homeostasis. B. megaterium ZS-3 increases the K+/Na+ ratio by directly limiting the accumulation of Na+, rather than by increasing the K+ content, in both the presence and absence of salt. Furthermore, B. megaterium ZS-3 determines the down-regulation of HKT1 and the up-regulation of NHX1 and AVP1 (a vacuolar H+-pyrophosphatase) pumping H+ into vesicles, which acidify to generate a H+ gradient across the membrane.
3.4.4. Hormonal Modulation
Plants have developed several strategies to cope with salt stress by optimizing the balance between growth and stress responses, and plenty of evidence suggests a critical role for phytohormones. Abscisic acid (ABA), salicylic acid (SA), and jasmonic acid (JA) are considered stress hormones, while auxins (IAA), cytokinins (CKs), and gibberellin (GB) are considered growth promoters [72]. Most studies on hormone modulation induced by PGPR are about ABA and its induced pathways. Indeed, among all phytohormones ABA is the main factor in the regulation of resistance to abiotic stresses in plants, which coordinates a variety of functions. Its activity depends on its concentration in the plant. At a normal level, ABA regulates various physiological processes such as stomatal opening, embryo morphogenesis, seed development, dormancy, and the synthesis of storage proteins and lipids [73][74], while at high concentrations, ABA inhibits plant growth. Under conditions of abiotic stress, such as drought and salt stresses, ABA biosynthesis is strongly induced, leading to an increase in its content in the plant, and determining stomatal closure and a change in gene expression, which may favor plant adaptation and survival [75].
The importance of ABA in this phenomenon is represented by the fact that in Arabidopsis seedlings, the genes modulated by ABA comprise about 10% of the genome, evenly divided between induced and repressed genes, i.e., two to six times more than those modulated by other plant hormones [76]. Most ABA-modulated genes encode proteins involved in stress tolerance, such as dehydrins and enzymes that detoxify ROS, as well as those involved in osmolyte metabolism, several transporters, transcription factors, protein kinases and phosphatases, and enzymes involved in phospholipid signaling [77]. Furthermore, ABA biosynthesis is also regulated by its end products since ABA negatively regulates its own accumulation by activating its catabolic enzymes [78].
During salt stress, many transcription factors (TFs) such as MYC, MYB, bZIP, MADS, and BHLB play a fundamental role in the response to stress. TFs generally act as key negative or positive regulators of gene expression. For instance, the regulation of MYB TFs in response to salt stress involves ABA. When plants grow under high salinity conditions, the ABA content increases significantly, initiating a cascade of salt stress response signals in the ABA-dependent pathway that leads to the up- or down-regulation of downstream response genes.
Ethylene is a key gaseous phytohormone with a wide range of biological activities that can affect plant growth and development. At high concentrations, it induces defoliation and other cellular processes that may lead to an inhibition of root and stem growth and premature senescence, reducing crop performance [79][80]. Under normal conditions, ethylene is produced, starting from 1-aminocyclopropane-1-carboxylate (ACC) through the action of the ACC synthetase enzyme. As a response to exposure to various environmental stresses such as drought, salt stress, cold, infections, heavy metals, and flooding, plants increase the production of ACC, resulting in a rise in ethylene concentration [81]. Auxins, such as IAA, control a wide range of processes in plant development and growth [82]. By increasing both the surface area and the length of the roots, IAA allows plants to have greater access to soil nutrients [83].
PGPR can modulate plant hormones to alleviate salt stress symptoms. Most PGPR can produce IAA, thus stimulating the development of secondary roots. Furthermore, by increasing the absorption of nutrients and water, IAA alleviates the effects of some abiotic stresses such as salinity and drought [84]. The production of the ACC-deaminase enzyme is another important physiological trait of PGPR that facilitates plant growth [85][86]. Indeed, under stressful conditions, when the level of ethylene in the plant might reach inhibitory levels, this enzyme supports plant growth by degrading ACC [26][87][88].
3.4.5. Nutrient Uptake
A high Na+ concentration induces nutritional disorders that reduce the activity of many essential nutrients in the soil, making them less available to plants. Salinity reduces the uptake and translocation of nitrogen (N), potassium (K+), phosphorous (P), calcium (Ca2+), and iron (Fe) [7][89][90]. Micronutrients may be less important for plants compared to N, K+, Ca2+, and P in terms of resistance to salinity [91]. The ability to fix nitrogen is widespread among prokaryotes [92]. An estimated 80% of the biological nitrogen fixation comes from symbiotic association, while the rest is provided by free-living (diazotrophs) or associative systems [93]. Non-symbiotic nitrogen-fixing rhizobacteria mainly belong to the Azoarcus, Azotobacter, Acetobacter, Azospirillum, Burkholderia, Diazotrophicus, Enterobacter, Gluconacetobacter, and Pseudomonas genera [79][94]. The inoculation of biological N2-fixing PGPR in crops promotes plant growth, reduces the need for pesticides, and restores nitrogen levels in agricultural soil under adverse environmental conditions such as salinity [95]. After nitrogen, P is the most important element in plant nutrition. It is involved in almost all major metabolic processes, such as signal transduction, respiration, photosynthesis, macromolecular biosynthesis, and energy transfer [96].
Fe is a key component of various metabolic pathways. More than 100 metabolic enzymes require iron as a cofactor and are essential for many plant processes, including photosynthesis, electron transport, oxidative phosphorylation, and hormone production [97]. Although it is the fourth most abundant element on earth, this huge amount of iron is not bioavailable for plants since free Fe(II) is rapidly oxidized to Fe(III), which is not assimilable due to its low solubility [98]. PGPR can produce siderophores, whose main function is to convert insoluble Fe to a form accessible to microorganisms [99]. Siderophores are also known to provide plants with iron and promote their growth [100].
Zn is one of the micronutrients required at a small concentration (5–100 mg kg−1) and is a co-factor and a metal activator of many enzymes [101]. Zn deficiency affects membrane integrity, and the synthesis of carbohydrates, auxins, and nucleotides [102].
Following the inoculation of Kosaconia radicincitans KR-17 in Raphanus sativus L. grown under saline conditions, Shahid et al. [103] found the content of N, P, K+, Ca2+, Mg, Zn, Fe, Cu, and Na+ to rise maximally compared to non-inoculated plants. In a study on C. quinoa, the inoculation of the Pseudomonas sp. M30-35 strain mitigated salt stress, maintaining P content and homeostasis [104]. The application of Priestia endophytica SK1 isolated from rhizospheric soil of fenugreek in Trigonella foenum-graecum induced nodule formation and was found to raise the content of N and P under 100 mM NaCl compared to control plants [105]. The inoculation of Glutamicibacter sp. and Pseudomonas sp. in Suaeda fruticose resulted in a reduction in Na+ and Cl− in the shoots of stressed plants. The inoculation of Pseudomonas sp. increased the K+ and Ca2+ content and improved the activities of urease, ß-glucosidase, and dehydrogenase compared to their corresponding non-inoculated stressed plants [106]. Comparative transcriptomic analyses between wheat roots inoculated with Arthrobacter nitroguajacolicus or not, revealed that under salt stress, the inoculation induces the up-regulation of various genes related to metal binding and involved in iron acquisition [107]
3.4.6. Biofilm Formation
Biofilms are extracellular matrices composed of proteins, nucleic acids, lipids, exopolysaccharides (EPS), and embedded microorganisms, which allow rhizosphere bacteria to adhere to the surface of plant roots [108]. The biofilms produced by PGPR also protect plants from stress conditions such as drought and salinity, as their components can co-ordinately function as osmoprotectors [109]. EPS is a natural mixture of high-molecular-weight polymers released by bacteria into the environment to mitigate physiological stress and plant environmental stresses such as salinity [110].
3.4.7. Volatile Organic Compounds (VOCs)
Volatile organic compounds (VOCs) are usually a mixture of metabolites produced by microorganisms during primary or secondary metabolism, characterized by their low molecular weight (>300 Da), lipophilic nature, and low boiling point. VOCs can travel from the point of production through soils, liquid, and atmosphere, which makes them ideal chemical messengers for intra- and inter-organismic communication [111][112]. The production of VOCs by microorganisms depends on the microbial species and can be affected by many environmental factors such as pH, temperature, humidity, etc. [113].
PGPR synthesize a broad range of VOCs, including aliphatic aldehydes, esters, alcohols, organic acids, ethers, ketones, sulfur-containing compounds, and hydrocarbons. Mainly known to be involved in biocontrol activity against plant phytopathogens or the stimulation of plant defense mechanisms, VOCs produced by PGPR also affect plant growth and abiotic stress tolerance [114].
4. Conclusions
In conclusion, PGPR have been shown to be effective in improving plant growth under salinity. Thanks to these bacteria’s ability to grow under salt stress conditions and their effects on plant growth, they are a very interesting, sustainable solution for improving crop productivity in saline soils. Although some PGPR have been associated with the modulation of genes belonging to different signal transduction pathways such as MYC, DREB2, and WRKY, extensive research remains to be conducted to understand the signaling pathways involved in plant and PGPR cross-talk, inducing the plant’s salt tolerance response.
References
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473.
- Smith, D.L.; Praslickova, D.; Ilangumaran, G. Inter-organismal signaling and management of the phytomicrobiome. Front. Plant Sci. 2015, 6, 722.
- Bringhurst, R.M.; Cardon, Z.G.; Gage, D.J. Galactosides in the rhizosphere: Utilization by Sinorhizobium meliloti and development of a biosensor. Proc. Natl. Acad. Sci. USA 2001, 98, 4540–4545.
- Hiltner, L. Über neuere erfahrungen und probleme auf dem debiete der bo denbakteriologie und unter besonderer berucksichtigung der grundund und brache. Zbl. Bakteriol. 1904, 2, 14–25.
- Gray, E.J.; Smith, D.L. Intracellular and extracellular PGPR: Commonalities and distinctions in the plant-bacterium signaling processes. Soil Biol. Biochem. 2005, 37, 395–412.
- Zhang, R.; Vivanco, J.M.; Shen, Q. The unseen rhizosphere root-soil-microbe interactions for crop production. Curr. Opin. Microbiol. 2017, 37, 8–14.
- Clarkson, D.T.; Marschner, H. Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: London, UK, 1995.
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant. Biol. 2013, 64, 807–838.
- Kumar, N.; Iyer-Pascuzzi, A.N. Shedding the last layer: Mechanisms of root cap cell release. Plants 2020, 9, 308.
- Upadhyay, S.K.; Srivastava, A.K.; Rajput, V.D.; Chauhan, P.K.; Bhojiya, A.A.; Jain, D.; Chaubey, G.; Dwivedi, P.; Sharma, B.; Minkina, T. Root exudates: Mechanistic insight of plant growth promoting rhizobacteria for sustainable crop production. Front. Microbiol. 2022, 13, 916488.
- Kawa, D.; Brady, S.M. Root cell types as an interface for biotic interactions. Trends Plant Sci. 2022, 27, 1173–1186.
- Ganesh, A.; Shukla, V.; Mohapatra, A.; George, A.P.; Bhukya, D.P.N.; Das, K.K.; Kola, V.S.R.; Suresh, A.; Ramireddy, E. Root cap to soil interface: A driving force toward plant adaptation and development. Plant Cell Physiol. 2022, 63, 1038–1051.
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 2019, 10, 157.
- Jones, P.; Garcia, B.J.; Furches, A.; Tuskan, G.A.; Jacobson, D. Plant host-associated mechanisms for microbial selection. Front. Plant Sci. 2019, 10, 862.
- Burdman, S.; Jurkevitch, E.; Okon, Y. Recent advances in the use of plant growth promoting rhizobacteria (PGPR) in agriculture. In Microbial Interactions in Agriculture and Forestry; Subba Rao, N.S., Dommergues, Y.R., Eds.; Science Publishers: Enfield, NH, USA, 2000; pp. 229–250.
- Kaymak, D.C. Potential of PGPR in agricultural innovations. In Plant Growth and Health Promoting Bacteria; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2010.
- Weller, D.M.; Thomashow, L.S. Current challenges in introducing beneficial microorganisms into the rhizosphere. In Molecular Ecology of Rhizosphere Microorganisms: Biotechnology and Release of GMOs; O’Gara, F., Dowling, D.N., Boesten, B., Eds.; VCH: New York, NY, USA, 1994; pp. 1–18.
- Soto, M.; Nogales, J.; Perez-Mendoza, D.; Gallegos, M.T.; Olivares, J.; Sanjuan, J. Pathogenic and mutualistic plant-bacteria interactions: Ever increasing similarities. Open Life Sci. 2011, 6, 911917.
- Santoyo, G.; Urtis-Flores, C.A.; Loeza-Lara, P.D.; Orozco-Mosqueda, M.D.C.; Glick, B.R. Rhizosphere colonization determinants by plant growth-promoting rhizobacteria (PGPR). Biology 2021, 10, 475.
- Kloepper, J.W.; Schroth, M.N. Plant growth-promoting rhizobacteria on radishes. In Proceedings of the 4th International Conference on Plant Pathogenic Bacteria, Angers, France, 27 August–2 September 1978; Gilbert-Clarey: Tours, France, 1978; pp. 879–882.
- Kloepper, J.W.; Schroth, M.N. Relationship of in vitro antibiosis of plant growth promoting rhizobacteria to plant growth and the displacement of root microflora. Phytopathology 1981, 71, 1020–1024.
- Antoun, H.; Kloepper, J.W. Plant growth promoting rhizobacteria. In Encyclopedia of Genetics; Brenner, S., Miller, J.H., Eds.; Academic: New York, NY, USA, 2001; pp. 1477–1480.
- Nakkeeran, S.; Fernando, W.G.D.; Siddiqui, Z.A. Plant Growth Promoting Rhizobacteria formulations and its scope in commercialization for the management of pests and diseases. In PGPR: Biocontrol and Biofertilization; Siddiqui, Z.A., Ed.; Springer: Dordrecht, The Netherlands, 2005; pp. 257–296.
- Martinez-Viveros, O.; Jorquera, M.A.; Crowley, D.E.; Gajardo, G.; Mora, M.L. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci. Plant. Nutr. 2010, 10, 293–319.
- Wang, E.T.; Martinez-Romero, E. Sesbania herbacea-Rhizobium huautlense nodulation in flooded soils and comparative characterization of S. herbacea-nodulating rhizobia in different environments. Microb. Ecol. 2000, 40, 25–32.
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39.
- Hayat, R.; Ali, S.; Amara, U. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598.
- Choudhary, D.K.; Sharma, K.P.; Gaur, R.K. Biotechnological perspectives of microbes in agro-ecosystems. Biotechnol. Lett. 2011, 33, 1905–1910.
- Bertola, M.; Ferrarini, A.; Visioli, G. Improvement of soil microbial diversity through sustainable agricultural practices and its evaluation by -Omics approaches: A perspective for the environment, food quality and human safety. Microorganisms 2021, 9, 1400.
- Giannelli, G.; Bisceglie, F.; Pelosi, G.; Bonati, B.; Cardarelli, M.; Antenozio, M.L.; Degola, F.; Visioli, G. Phyto-beneficial traits of rhizosphere bacteria: In vitro exploration of plant growth promoting and phytopathogen biocontrol ability of selected strains isolated from harsh environments. Plants 2022, 11, 230.
- Kumar, P.; Sharma, P.K. Soil salinity and food security in India. Front. Sustain. Food Syst. 2020, 4, 533781.
- Omuto, C.T.; Vargas, R.R.; El Mobarak, A.M.; Mohamed, N.; Viatkin, K.; Yigini, Y. Mapping of Salt-Affected Soils: Technical Manual; FAO: Rome, Italy, 2020.
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131.
- Maathuis, F.J.M. Sodium in plants: Perception, signalling, and regulation of sodium fluxes. J. Exp. Bot. 2014, 65, 849–858.
- FAO. Global Map of Salt Affected Soils. 2021. Available online: https://www.fao.org/soils-portal/data-hub/soil-maps-and-databases/global-map-of-salt-affected-soils/en/ (accessed on 17 January 2022).
- Carillo, P.; Annunziata, M.G.; Pontecorvo, G.; Fuggi, A.; Woodrow, P. Salinity stress and salt tolerance. In Abiotic Stress in Plants—Mechanisms and Adaptations; Shanker, A., Venkateswarlu, B., Eds.; IntechOpen: London, UK, 2011.
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681.
- İbrahimova, U.; Kumari, P.; Yadav, S.; Rastogi, A.; Antala, M.; Suleymanova, Z.; Zivcak, M.; Tahjib-Ul-Arif, M.; Hussain, S.; Abdelhamid, M.; et al. Progress in understanding salt stress response in plants using biotechnological tools. J. Biotechnol. 2021, 329, 180–191.
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genomics 2014, 2014, 701596.
- Ondrasek, G.; Rathod, S.; Manohara, K.K.; Gireesh, C.; Anantha, M.S.; Sakhare, A.S.; Parmar, B.; Yadav, B.K.; Bandumula, N.; Raihan, F.; et al. Salt stress in plants and mitigation approaches. Plants 2022, 11, 717.
- Kumar, A.; Singh, S.; Mukherjee, A.; Rastogi, R.P.; Verma, J.P. Salt-tolerant plant growth-promoting Bacillus pumilus strain JPVS11 to enhance plant growth attributes of rice and improve soil health under salinity stress. Microbiol. Res. 2021, 242, 126616.
- Nadeem, S.M.; Ahmad, M.; Naveed, M.; Imran, M.; Zahir, Z.A.; Crowley, D.E. Relationship between in vitro characterization and comparative efficacy of plant growth-promoting rhizobacteria for improving cucumber salt tolerance. Arch. Microbiol. 2016, 198, 379–387.
- Fouda, A.; Hassan, S.; Eid, A.; Ewais, E. The interaction between plants and bacterial endophytes under salinity stress. In Endophytes and Secondary Metabolites; Jha, S., Ed.; Reference Series in Phytochemistry; Springer: Cham, Switzerland, 2019; pp. 1–17.
- Bhat, M.A.; Kumar, V.; Bhat, M.A.; Wani, I.A.; Dar, F.L.; Farooq, I.; Bhatti, F.; Koser, R.; Rahman, S.; Jan, A.T. Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (PGPR) with plant roots toward enhancing plant productivity by alleviating salinity stress. Front. Microbiol. 2020, 11, 1952.
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433.
- Ilangumaran, G.; Smith, D.L. Plant growth promoting rhizobacteria in amelioration of salinity stress: A systems biology perspective. Front. Plant Sci. 2017, 8, 1768.
- Kumawat, K.C.; Sharma, B.; Nagpal, S.; Kumar, A.; Tiwari, S.; Nair, R.M. Plant growth-promoting rhizobacteria: Salt stress alleviators to improve crop productivity for sustainable agriculture development. Front. Plant Sci. 2023, 13, 1101862.
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673.
- Raven, J.A. Regulation of Ph and generation of osmolarity in vascular plants—A cost-benefit analysis in relation to efficiency of use of energy, nitrogen and water. New Phytol. 1985, 101, 25–77.
- Asensi-Fabado, M.A.; Amtmann, A.; Perrella, G. Plant responses to abiotic stress: The chromatin context of transcriptional regulation. Biochim. Biophys. Acta—Gene Regul. Mech. 2017, 1860, 106–122.
- Cui, J.; Sun, T.; Li, S.; Xie, Y.; Song, X.; Wang, F.; Chen, L. Improved salt tolerance and metabolomics analysis of Synechococcus elongatus UTEX 2973 by overexpressing Mrp Antiporters. Front. Bioeng. Biotechnol. 2020, 8, 500.
- Reza Amirjani, M. Effect of salinity stress on growth, sugar content, pigments and enzyme activity of rice. Int. J. Bot. 2011, 7, 73–81. Available online: https://scialert.net/abstract/?doi=ijb.2011.73.81 (accessed on 11 June 2020).
- Jogawat, A. Osmolytes and their role in abiotic stress tolerance in plants. In Molecular Plant Abiotic Stress; Roychoudhury, A., Tripathi, D., Eds.; Wiley: Hoboken, NJ, USA, 2019.
- De Zelicourt, A.; Colcombet, J.; Hirt, H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016, 21, 677–685.
- Kim, S.H.; Woo, D.H.; Kim, J.M.; Lee, S.Y.; Chung, W.S.; Moon, Y.H. Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem. Biophys. Res. Commun. 2011, 412, 150–154.
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385.
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132.
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593.
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399.
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18.
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotech. 2005, 16, 123–132.
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804.
- Damodaran, T.; Rai, R.B.; Jha, S.K.; Kannan, R.; Pandey, B.K.; Sah, V.; Mishra, V.K.; Sharma, D.K. Rhizosphere and endophytic bacteria for induction of salt tolerance in gladiolus grown in sodic soils. J. Plant Interact. 2014, 9, 577–584.
- Etesami, H.; Beattie, G.A. Plant-Microbe interactions in adaptation of agricultural crops to abiotic stress conditions. In Probiotics and Plant Health; Kumar, V., Kumar, M., Sharma, S., Prasad, R., Eds.; Springer: Singapore, 2017; pp. 163–200.
- Kim, K.; Jang, Y.J.; Lee, S.M.; Oh, B.T.; Chae, J.C.; Lee, K.J. Alleviation of salt stress by Enterobacter sp. EJ01 in tomato and Arabidopsis is accompanied by upregulation of conserved salinity responsive factors in plants. Mol. Cells 2014, 37, 109–117.
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J.M. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40, 326–345.
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663.
- Singh, A.; Roychoudhury, A. Gene regulation at transcriptional and post-transcriptional levels to combat salt stress in plants. Physiol. Plant. 2021, 173, 1556–1572.
- Guo, Y.; Qiu, Q.S.; Quintero, F.J.; Pardo, J.M.; Ohta, M.; Zhang, C.; Schumaker, K.S.; Zhu, J.K. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 2004, 16, 435–449.
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901.
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477.
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130.
- Finkelstein, R.R.; Rock, C.D. Abscisic acid biosynthesis and response. In The Arabidopsis Book; Somerville, C.R., Meyerowitz, E.M., Eds.; American Society of Plant Biologists: Rockville, MD, USA, 2002.
- Sreenivasulu, N.; Harshavardhan, V.T.; Govind, G.; Seiler, C.; Kohli, A. Contrapuntal role of ABA: Does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 2012, 506, 265–273.
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571.
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475.
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679.
- Cutler, A.J.; Krochko, J.E. Formation and breakdown of ABA. Trends Plant Sci. 1999, 4, 472–478.
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350.
- Li, Q.; Saleh-Lakha, S.; Glick, B.R. The effect of native and ACC deaminase containing Azospirillum brasilense Cdl843 on the rooting of carnation cuttings. Can. J. Microbiol. 2005, 51, 511–514.
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401.
- Bhardwaj, D.; Ansari, M.W.; Sahoo, R.K.; Tuteja, N. Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb. Cell Fact. 2014, 13, 66.
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448.
- Etesami, H.; Beattie, G.A. Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Front. Microbiol. 2018, 9, 148.
- Etesami, H.; Alikhani, H.A.; Mirseyed Hosseini, H. Indole-3-acetic Acid and 1-Aminocyclopropane-1-Carboxylate Deaminase: Bacterial traits required in rhizosphere, rhizoplane and/or endophytic competence by beneficial bacteria. In Bacterial Metabolites in Sustainable Agroecosystem; Maheshwari, D., Ed.; Sustainable Development and Biodiversity; Springer: Cham, Switzerland, 2015; Volume 12.
- Glick, B.R. Bacterial ACC deaminase and the alleviation of plant stress. Adv. Appl. Microbiol. 2004, 56, 291–312.
- Gamalero, E.; Glick, B.R. Bacterial modulation of plant ethylene levels. Plant Physiol. 2015, 169, 13–22.
- Glick, B.R.; Cheng, Z.; Czarny, J.; Duan, J. Promotion of plant growth by ACC deaminase-producing soil bacteria. Eur. J. Plant Pathol. 2007, 119, 329–339.
- Ben Abdallah, H.; Mai, H.J.; Alvarez-Fernandez, A.; Abadia, J.; Bauer, P. Natural variation reveals contrasting abilities to cope with alkaline and saline soil among different Medicago truncatula genotypes. Plant Soil 2017, 418, 45–60.
- Läuchli, A.; Grattan, S. Plant growth and development under salinity stress. In Advances in Molecular Breeding toward Drought and Salt Tolerant Crops; Jenks, M.A., Hasegawa, P.M., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2007.
- Hu, Y.; Schmidhalter, U. Drought and salinity: A comparison of their effects on mineral nutrition of plants. J. Plant Nutr. Soil Sci. 2005, 168, 541–549.
- Singh, M.; Singh, D.; Gupta, A.; Pandey, K.D.; Singh, P.K.; Kumar, A. Plant Growth Promoting Rhizobacteria: Application in Biofertilizers and Biocontrol of Phytopathogens. In PGPR Amelioration in Sustainable Agriculture; Singh, A.K., Kumar, A., Singh, P.K., Eds.; Woodhead Publishing: Sawston, UK, 2019; pp. 41–66.
- Tilak, K.V.B.R.; Ranganayaki, N.; Pal, K.K.; De, R.; Saxena, A.K.; Nautiyal, C.S.; Mittal, S.; Tripathi, A.K.; Johri, B.N. Diversity of plant growth and soil health supporting bacteria. Curr. Sci. 2005, 89, 136–150. Available online: http://www.jstor.org/stable/24110439 (accessed on 11 June 2020).
- Vessey, J.K. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 2003, 255, 571–586.
- Damam, M.; Kaloori, K.; Gaddam, B.; Kausar, R. Plant growth promoting substances (phytohormones) produced by rhizobacterial strains isolated from the rhizosphere of medicinal plants. Int. J. Pharm. Sci. Rev. Res. 2016, 37, 130–136.
- Khan, M.S.; Zaidi, A.; Ahemad, M.; Oves, M.; Wani, P.A. Plant growth promotion by phosphate solubilizing fungi—Current perspective. Arch. Agron. Soil Sci. 2010, 56, 73–98.
- Timofeeva, A.M.; Galyamova, M.R.; Sedykh, S.E. Bacterial siderophores: Classification, biosynthesis, perspectives of use in agriculture. Plants 2022, 11, 3065.
- Ammari, T.; Mengel, K. Total soluble Fe in soil solutions of chemically different soils. Geoderma 2006, 136, 876–885.
- Dertz, E.A.; Xu, J.; Stintzi, A.; Raymond, K.N. Bacillibactin-mediated iron transport in Bacillus subtilis. J. Am. Chem. Soc. 2006, 11, 22–23.
- Scavino, A.F.; Pedraza, R.O. The role of siderophores in plant growth-promoting bacteria. In Bacteria in Agrobiology: Crop Productivity; Maheshwari, D., Saraf, M., Aeron, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2013.
- Parisi, B.; Vallee, B.L. Metal enzyme complexes activated by zinc. J. Biol. Chem. 1969, 179, 803–807.
- Singh, B.P.; Natesan, S.; Singh, B.; Usha, K. Improving zinc efficiency of cereals under zinc deficiency. Curr. Sci. 2005, 88, 36–44.
- Shahid, M.; Al-Khattaf, F.S.; Danish, M.; Zeyad, M.T.; Atef Hatamleh, A.; Mohamed, A.; Ali, S. PGPR Kosakonia Radicincitans KR-17 increases the salt tolerance of radish by regulating ion-homeostasis, photosynthetic molecules, redox potential, and stressor metabolites. Front. Plant Sci. 2022, 13, 919696.
- Cai, D.; Xu, Y.; Zhao, F.; Zhang, Y.; Duan, H.; Guo, X. Improved salt tolerance of Chenopodium quinoa Willd. Contributed by Pseudomonas sp. Strain M30-35. PeerJ 2021, 9, e10702.
- Sharma, K.; Sharma, S.; Vaishnav, A.; Jain, R.; Singh, D.; Singh, H.B.; Goel, A.; Singh, S. Salt-tolerant PGPR strain Priestia endophytica SK1 promotes fenugreek growth under salt stress by inducing nitrogen assimilation and secondary metabolites. J. Appl. Microbiol. 2022, 133, 2802–2813.
- Hidri, R.; Mahmoud, O.M.; Zorrig, W.; Mahmoudi, H.; Smaoui, A.; Abdelly, C.; Azcon, R.; Debez, A. Plant growth-promoting rhizobacteria alleviate high salinity impact on the halophyte Suaeda fruticose by modulating antioxidant defense and soil biological activity. Front. Plant Sci. 2022, 13, 821475.
- Safdarian, M.; Askari, H.; Shariati, J.V.; Nematzadeh, G. Transcriptional responses of wheat roots inoculated with Arthrobacter nitroguajacolicus to salt stress. Sci. Rep. 2019, 9, 1792.
- Danhorn, T.; Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422.
- Rojas-Solis, D.; Vences-Guzmán, M.Á.; Sohlenkamp, C.; Santoyo, G. Antifungal and plant growth–promoting Bacillus under saline stress modify their membrane composition. J. Soil Sci. Plant Nutr. 2020, 20, 1549–1559.
- Morcillo, R.J.L.; Manzanera, M. The effects of plant-associated bacterial exopolysaccharides on plant abiotic stress tolerance. Metabolites 2021, 11, 337.
- Kanchiswamy, C.N.; Malnoy, M.; Maffei, M.E. Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front. Plant Sci. 2015, 6, 151.
- Maffei, M.E.; Gertsch, J.; Appendino, G. Plant volatiles production, function and pharmacology. Nat. Prod. Rep. 2011, 28, 1359–1380.
- Korpi, A.; Järnberg, J.; Pasanen, A.L. Microbial volatile organic compounds. Crit. Rev. Toxicol. 2009, 39, 139–193.
- Bhattacharyya, D.; Lee, Y.H. A cocktail of volatile compounds emitted from Alcaligenes faecalis JBCS1294 induces salt tolerance in Arabidopsis thaliana by modulating hormonal pathways and ion transporters. J. Plant Physiol. 2017, 214, 64–73.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
2 times
(View History)
Update Date:
17 Aug 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




