
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Olga Senko | -- | 4313 | 2023-07-21 11:50:07 | | | |
| 2 | Dean Liu | -14 word(s) | 4299 | 2023-07-24 03:24:32 | | |
Video Upload Options
Combination of metals and enzymes as effective antifungal agents is currently being conducted due to the growing antifungal resistance problem. Metals are attracting special attention due to the wide variety of ligands that can be used for them, including chemically synthesized and naturally obtained variants as a result of the so-called “green synthesis”. The main mechanism of the antifungal action of metals is the triggering of the generation and accumulation of reactive oxygen species (ROS). Further action of ROS on various biomolecules is nonspecific. Various hydrolytic enzymes exhibit antifungal properties by affecting the structural elements of fungal cells (cell walls, membranes), fungal quorum sensing molecules, fungal own protective agents (mycotoxins and antibiotics), and proteins responsible for the adhesion and formation of stable, highly concentrated populations in the form of biofilms.
1. Introduction
2. Antifungal Agents Based on Metal Nanoparticles, Metal–Organic Frameworks and Their Composites
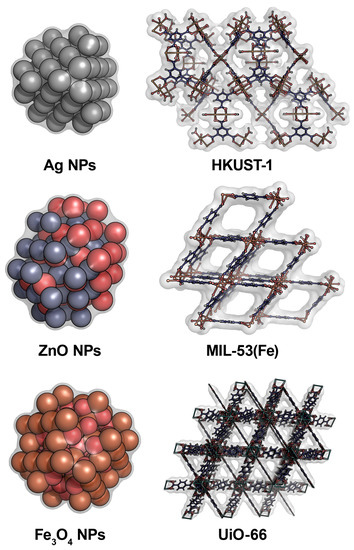
| Antifungal Agent [Reference] | Target of Action | Antifungal Activity | Efficiency of Antifungal Action |
|---|---|---|---|
| ZrO2-Ag2O (14–42 nm) [23] | Candida albicans, C. dubliniensis, C. glabrata, C. tropicalis |
The growth rate inhibition | 89–97% inhibition |
| WS2/ZnO nano-hybrids [24] | C. albicans | Inhibition of biofilm formation | 91% inhibition |
| CuO@C (36–123 nm) [25] | Alternaria alternata, Fusarium oxysporum, Penicillium digitatum, Rhizopus oryzae |
Inhibition of the hydrolytic activity of fungal enzymes used by them for their own metabolism | Inhibition (100 μg/mL) of cellulases and amylases secreted by fungi: 38% and 42% for A. alternata, 39% and 45% for F. oxysporum, 24% and 67% for P. digitatum, and 20% and 24%for R. oryzae, respectively |
| ZnO NPs [26] | C. albicans, Aspergillus niger |
Inhibition of growth | Large enough zone of growth absence (8-9 mm) |
| ZnO NPs (20–45 nm) [27] | Erythricium salmonicolor | Notable thinning of the hyphae and cell walls, liquefaction of the cytoplasmic content with decrease in presence of a number of vacuoles | Significant inhibition (9–12 mmol/L) of cell growth |
| ZnO–TiO2 NPs (8–33 nm) [28] | A. flavus | High level of ROS production and oxidative stress induction. Treated objects have a lower count of spores and damaged tubular filaments and noticeably thinner hyphae compared to the untreated fungi | Fungicidal inhibition (150 μg/mL) zone is 100 % |
| ZnO NPs (40–50 nm) [29] | C. albicans | High level of ROS production | MIC = 32–64 μg/mL MFC = 128–512 mg/mL |
| Fe2O3 NPs (10–30 nm) [30] | Trichothecium roseum, Cladosporium herbarum, P. chrysogenum, A. alternata, A. niger |
Inhibition of spore germination | MIC = 0.063–0.016 mg/mL |
| Fe3O4 NPs (70 nm) [31] | C. albicans | Inhibition of cell growth and biofilm formation | MIC = 100 ppm MFC = 200 ppm |
| Cu-BTC (10–20 µm) [32] | C. albicans, A. niger, A. oryzae, F. oxysporum |
ROS producing, the damage of the cell membrane | Inhibition of C. albicans colonies is 96% by 300 ppm and up to 100% by 500 ppm. Inhibition growth of F. oxysporum and A. oryzae is 30% with 500 ppm. No significant effect on the A. niger growth. |
| HKUST-1 or HKUST-1 NPs (doped with NPs of Cu(I)) (49–51 nm) [33] | A. niger, F. solani, P. chrysogenum |
Appearance of Cu+2 inhibiting of cell growth | 100% growth inhibition of F. solani by 750–1000 ppm and P. chrysogenum by 1000 ppm; for A. niger—no inhibition |
| [Cu2(Glt)2(LIGAND)] (H2O) [34] | C. albicans, A. niger spores |
The apoptosis-like fungal cell death, ROS production | 50–70% death of C. albicans and 50–80% germination inhibition of A. niger at 2 mg/mL of the MOFs |
| MIL-53(Fe) and Ag@MIL-53(Fe) composite [35] | A. flavus | Inhibition of cell growth | MIC = 40 μg/mL for the MIL-53(Fe); MIC = 15 μg/mL for the Ag@MIL-53(Fe) |
| MOF on the basis of Ce and 4,4′,4″-nitrilotribenzoic acid [11] | A. flavus, A. niger, Aspergillus terreus, C. albicans, Rhodotorula glutinis |
Enzyme-like activity: catalase, superoxide dismutase, and peroxidase | Inhibition efficiency of 93.3–99.3% based on the colony-forming unit method |
| TiO2 co-doped with nitrogen and fluorine (200–300 nm) [12] | F. oxysporum | Peroxidase-like activity, production of ROS under light irradiation | 100% inhibition of fungal growth |
| Fe3O4@MoS2-Ag (~428.9 nm) [36] | C. albicans | Peroxidase-like activity | 80% damage of cell membranes |
| CoZnO/MoS2 nanocomposite [37] | A. flavus | Peroxidase-like activity under light irradiation | MIC = 1.8 mg/mL |
3. Enzymes as Antifungal Agents
3.1. Antifungal Enzymes Using Cell Structural Components of Fungi as Substrates
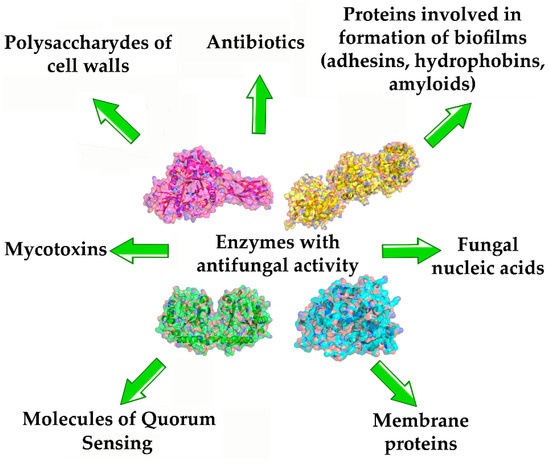
3.2. Enzymes Hydrolyzing Fungal Proteins with Amyloid Characteristics
3.3. Enzymes Hydrolyzing Mycotoxins, Antibiotics, and QS Molecules (QSMs) of Fungi
4. Combination of Antifungal Enzymes and Metal-Nanoparticles
References
- Fisher, M.C.; Gurr, S.J.; Cuomo, C.A.; Blehert, D.S.; Jin, H.; Stukenbrock, E.H.; Stajich, J.E.; Regine Kahmann, R.; Boone, C.; Denning, D.W.; et al. Threats posed by the fungal kingdom to humans, wildlife, and agriculture. MBio 2020, 11, e00449-20.
- Garg, D.; Muthu, V.; Sehgal, I.S.; Ramachandran, R.; Kaur, H.; Bhalla, A.; Puri, G.D.; Chakrabarti, A.; Agarwal, R. Coronavirus disease (COVID-19) associated mucormycosis (CAM): Case report and systematic review of literature. Mycopathologia 2021, 186, 289–298.
- Raut, A.; Huy, N.T. Rising incidence of mucormycosis in patients with COVID-19: Another challenge for India amidst the second wave? Lancet Respir. Med. 2021, 9, e77.
- World Health Organization. WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; World Health Organization: Geneva, Switzerland, 2022; p. 48. Available online: https://www.who.int/publications/i/item/9789240060241 (accessed on 29 May 2023).
- Robbins, N.; Caplan, T.; Cowen, L.E. Molecular evolution of antifungal drug resistance. Annu. Rev. Microbiol. 2017, 71, 753–775.
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018, 360, 739–742.
- Rabaan, A.A.; Sulaiman, T.; Al-Ahmed, S.H.; Buhaliqah, Z.A.; Buhaliqah, A.A.; AlYuosof, B.; Alfaresi, M.; Al Fares, M.A.; Alwarthan, S.; Alkathlan, M.S.; et al. Potential strategies to control the risk of antifungal resistance in humans: A comprehensive review. Antibiotics 2023, 12, 608.
- WHO. WHO Releases First-Ever List of HEALTH-Threatening Fungi; World Health Organization: Geneva, Switzerland, 2022; Available online: https://www.who.int/news/item/25-10-2022-who-releases-first-ever-list-of-health-threatening-fungi (accessed on 30 May 2023).
- Cruz-Luna, A.R.; Cruz-Martínez, H.; Vásquez-López, A.; Medina, D.I. Metal nanoparticles as novel antifungal agents for sustainable agriculture: Current advances and future directions. J. Fungi 2021, 7, 1033.
- Dananjaya, S.H.S.; Thao, N.T.; Wijerathna, H.M.S.M.; Lee, J.; Edussuriya, M.; Choi, D.; Kumar, R.S. In vitro and in vivo anticandidal efficacy of green synthesized gold nanoparticles using Spirulina maxima polysaccharide. Process Biochem. 2020, 92, 138–148.
- Abdelhamid, H.N.; Mahmoud, G.A.E.; Sharmouk, W. A cerium-based MOFzyme with multi-enzyme-like activity for the disruption and inhibition of fungal recolonization. J. Mater. Chem. B 2020, 8, 7548–7556.
- Mukherjee, K.; Acharya, K.; Biswas, A.; Jana, N.R. TiO2 nanoparticles co-doped with nitrogen and fluorine as visible-light-activated antifungal agents. ACS Appl. Nano Mater. 2020, 3, 2016–2025.
- Wen, H.; Shi, H.; Jiang, N.; Qiu, J.; Lin, F.; Kou, Y. Antifungal mechanisms of silver nanoparticles on mycotoxin producing rice false smut fungus. Iscience 2023, 26, 105763.
- Malik, M.A.; Batterjee, M.G.; Kamli, M.R.; Alzahrani, K.A.; Danish, E.Y.; Nabi, A. Polyphenol-capped biogenic synthesis of noble metallic silver nanoparticles for antifungal activity against Candida auris. J. Fungi 2022, 8, 639.
- Soleimani, P.; Mehrvar, A.; Michaud, J.P.; Vaez, N. Optimization of silver nanoparticle biosynthesis by entomopathogenic fungi and assays of their antimicrobial and antifungal properties. J. Invertebr. Pathol. 2022, 190, 107749.
- Jamdagni, P.; Khatri, P.; Rana, J.S. Green synthesis of zinc oxide nanoparticles using flower extract of Nyctanthes arbor-tristis and their antifungal activity. J. King Saud Univ. Sci. 2018, 30, 168–175.
- Jamdagni, P.; Rana, J.S.; Khatri, P.; Nehra, K. Comparative account of antifungal activity of green and chemically synthesized zinc oxide nanoparticles in combination with agricultural fungicides. Int. J. Nano Dimens. 2018, 9, 198–208.
- Zhou, L.; Zhao, X.; Li, M.; Lu, Y.; Ai, C.; Jiang, C.; Liu, Y.; Pan, Z.; Shi, J. Antifungal activity of silver nanoparticles synthesized by iturin against Candida albicans in vitro and in vivo. Appl. Microbiol. Biotechnol. 2021, 105, 3759–3770.
- Shamraychuk, I.L.; Belyakova, G.A.; Eremina, I.M.; Kurakov, A.V.; Belozersky, M.A.; Dunaevsky, Y.E. Fungal proteolytic enzymes and their inhibitors as perspective biocides with antifungal action. Mosc. Univ. Biol. Sci. Bull. 2020, 75, 97–103.
- Padder, S.A.; Prasad, R.; Shah, A.H. Quorum sensing: A less known mode of communication among fungi. Microbiol. Res. 2018, 210, 51–58.
- Baier, F.; Tokuriki, N. Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J. Mol. Biol. 2014, 426, 2442–2456.
- Lyagin, I.; Efremenko, E. Enzymes for detoxification of various mycotoxins: Origins and mechanisms of catalytic action. Molecules 2019, 24, 2362.
- Ayanwale, A.P.; Estrada-Capetillo, B.L.; Reyes-López, S.Y. Evaluation of antifungal activity by mixed oxide metallic nanocomposite against Candida spp. Processes 2021, 9, 773.
- Bhatt, V.K.; Patel, M.; Pataniya, P.M.; Iyer, B.D.; Sumesh, C.K.; Late, D.J. Enhanced antifungal activity of WS2/ZnO nanohybrid against Candida albicans. ACS Biomater. Sci. Eng. 2020, 6, 6069–6075.
- Abdelhamid, H.N.; Mahmoud, G.A.E. Antifungal and nanozyme activities of metal–organic framework-derived Appl. Organomet. Chem. 2023, 37, e7011.
- Pillai, A.M.; Sivasankarapillai, V.S.; Rahdar, A.; Joseph, J.; Sadeghfar, F.; Rajesh, K.; Kyzas, G.Z. Green synthesis and characterization of zinc oxide nanoparticles with antibacterial and antifungal activity. J. Mol. Struct. 2020, 1211, 128107.
- Arciniegas-Grijalba, P.A.; Patiño-Portela, M.C.; Mosquera-Sánchez, L.P.; Guerrero-Vargas, J.A.; Rodríguez-Páez, J.E. ZnO nanoparticles (ZnO-NPs) and their antifungal activity against coffee fungus Erythricium salmonicolor. Appl. Nanosci. 2017, 7, 225–241.
- Ilkhechi, N.N.; Mozammel, M.; Khosroushahi, A.Y. Antifungal effects of ZnO, TiO2 and ZnO-TiO2 nanostructures on Aspergillus flavus. Pestic. Biochem. Phys. 2021, 176, 104869.
- Miri, A.; Khatami, M.; Ebrahimy, O.; Sarani, M. Cytotoxic and antifungal studies of biosynthesized zinc oxide nanoparticles using extract of Prosopis farcta fruit. Green Chem. Lett. Rev. 2020, 13, 27–33.
- Parveen, S.; Wani, A.H.; Shah, M.A.; Devi, H.S.; Bhat, M.Y.; Koka, J.A. Preparation, characterization and antifungal activity of iron oxide nanoparticles. Microb. Pathog. 2018, 115, 287–292.
- Golipour, F.; Habibipour, R.; Moradihaghgou, L. Investigating effects of superparamagnetic iron oxide nanoparticles on Candida albicans biofilm formation. Med. Lab. J. 2019, 13, 44–50.
- Bouson, S.; Krittayavathananon, A.; Phattharasupakun, N.; Siwayaprahm, P.; Sawangphruk, M. Antifungal activity of water-stable copper-containing metal-organic frameworks. R. Soc. Open Sci. 2017, 4, 170654.
- Celis-Arias, V.; Loera-Serna, S.; Beltrán, H.I.; Álvarez-Zeferino, J.C.; Garrido, E.; Ruiz-Ramos, R. The fungicide effect of HKUST-1 on Aspergillus niger, Fusarium solani and Penicillium chrysogenum. New J. Chem. 2018, 42, 5570–5579.
- Veerana, M.; Kim, H.C.; Mitra, S.; Adhikari, B.C.; Park, G.; Huh, S.; Kim, S.; Kim, Y. Analysis of the effects of Cu-MOFs on fungal cell inactivation. RSC Adv. 2021, 11, 1057–1065.
- Tella, A.C.; Okoro, H.K.; Sokoya, S.O.; Adimula, V.O.; Olatunji, S.O.; Zvinowanda, C.; Ngila, J.C.; Shaibu, R.O.; Adeyemi, O.G. Synthesis, characterization and antifungal activity of Fe(III)metal–organic framework and its nano-composite. Chem. Afr. 2020, 3, 119–126.
- Wei, F.; Cui, X.; Wang, Z.; Dong, C.; Li, J.; Han, X. Recoverable peroxidase-like Fe3O4@MoS2-Ag nanozyme with enhanced antibacterial ability. Chem. Eng. J. 2021, 408, 127240.
- Liu, J.; Cheng, W.; Wang, Y.; Fan, X.; Shen, J.; Liu, H.; Wang, A.; Hui, A.; Nichols, F.; Chen, S. Cobalt-doped zinc oxide nanoparticle–MoS2 nanosheet composites as broad-spectrum bactericidal agents. ACS Appl. Nano Mater. 2021, 4, 4361–4370.
- Gow, N.A.R.; Latge, J.P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 10–128.
- Kühbacher, A.; Burger-Kentischer, A.; Rupp, S. Interaction of Candida species with the skin. Microorganisms 2017, 5, 32.
- Lyagin, I.; Stepanov, N.; Maslova, O.; Senko, O.; Aslanli, A.; Efremenko, E. Not a mistake but a feature: Promiscuous activity of enzymes meeting mycotoxins. Catalysts 2022, 12, 1095.
- Li, C.; Li, X.; Bai, C.; Zhang, Y.; Wang, Z. A chitinase with antifungal activity from naked oat (Avena chinensis) seeds. J. Food Biochem. 2019, 43, e12713.
- Dikbaş, N.; Uçar, S.; Tozlu, E.; Kotan, M.S.; ·Kotan, R. Antifungal activity of partially purified bacterial chitinase against Alternaria alternata. Erwerbs-Obstbau 2022.
- Zhang, W.; Ma, J.; Yan, Q.; Jiang, Z.; Yang, S. Biochemical characterization of a novel acidic chitinase with antifungal activity from Paenibacillus xylanexedens Z2–4. Int. J. Biol. Macromol. 2021, 182, 1528–1536.
- Rajninec, M.; Jopcik, M.; Danchenko, M.; Libantova, J. Biochemical and antifungal characteristics of recombinant class I chitinase from Drosera rotundifolia. Int. J. Biol. Macromol. 2020, 161, 854–863.
- Wang, N.-N.; Gao, K.-Y.; Han, N.; Tian, R.-Z.; Zhang, J.-L.; Yan, X.; Huang, L.-L. ChbB increases antifungal activity of Bacillus amyloliquefaciens against Valsa mali and shows synergistic action with bacterial chitinases. Biol. Control 2020, 142, 104150.
- Li, Q.; Hou, Z.; Zhou, D.; Jia, M.; Lu, S.; Yu, J. Antifungal activity and possible mechanism of Bacillus amyloliquefaciens FX2 against the postharvest apple ring rot pathogen. Phytopathology 2022, 112, 2486–2494.
- Lu, Y.; Wang, N.; He, J.; Li, Y.; Gao, X.; Huang, L.; Yan, X. Expression and characterization of a novel chitinase with antifungal activity from a rare actinomycete Saccharothrix yanglingensis Hhs.015. Protein Expr. Purif. 2018, 143, 45–51.
- Brzezinska, M.S.; Jankiewicz, U.; Kalwasinska, A.; Swiatczak, J.; Zero, K. Characterization of chitinase from Streptomyces luridiscabiei U05 and its antagonist potential against fungal plant pathogens. J. Phytopathol. 2019, 167, 404–412.
- Le, B.; Yang, S.H. Characterization of a chitinase from Salinivibrio sp. BAO-1801 as an antifungal activity and a biocatalyst for producing chitobiose. J. Basic Microbiol. 2018, 58, 848–856.
- Li, Z.; Xia, C.; Wang, Y.; Li, X.; Qiao, Y.; Li, C.; Zhou, J.; Zhang, L.; Ye, X.; Huang, Y.; et al. Identification of an endo-chitinase from Corallococcus sp. EGB and evaluation of its antifungal properties. Int. J. Biol. Macromol. 2019, 132, 1235–1243.
- Moon, C.; Seo, D.J.; Song, Y.S.; Hong, S.H.; Choi, S.H.; Jung, W.J. Antifungal activity and patterns of N-acetyl-chitooligosaccharide degradation via chitinase produced from Serratia marcescens PRNK-1. Microb. Pathog. 2017, 113, 218–224.
- Deng, J.-J.; Shi, D.; Mao, H.-H.; Li, Z.-W.; Liang, S.; Ke, Y.; Luo, X.-C. Heterologous expression and characterization of an antifungal chitinase (Chit46) from Trichoderma harzianum GIM 3.442 and its application in colloidal chitin conversion. Int. J. Biol. Macromol. 2019, 134, 113–121.
- Yilmaz, G.; Cadirci, B. Comparison of in vitro antifungal activity methods using Aeromonas sp. BHC02 chitinase, whose physicochemical properties were determined as antifungal agent candidate. Res. Sq. 2022.
- Sinitsyna, O.A.; Rubtsova, E.A.; Sinelnikov, I.G.; Osipov, D.O.; Rozhkova, A.M.; Matys, V.Y.; Bubnova, T.V.; Nemashkalov, V.A.; Sereda, A.S.; Tcsherbakova, L.A.; et al. Creation of chitinase producer and disruption of micromycete cell wall with the obtained enzyme preparation. Biochemistry 2020, 85, 717–724.
- Sachivkina, N.; Lenchenko, E.; Blumenkrants, D.; Ibragimova, A.; Bazarkina, O. Effects of farnesol and lyticase on the formation of Candida albicans biofilm. Vet. World 2020, 13, 1030–1036.
- Ling, L.; Cheng, W.; Jiang, K.; Jiao, Z.; Luo, H.; Yang, C.; Pang, M.; Lu, L. The antifungal activity of a serine protease and the enzyme production of characteristics of Bacillus licheniformis TG116. Arch. Microbiol. 2022, 204, 601.
- Deng, J.-J.; Huang, W.Q.; Li, Z.-W.; Lu, D.-L.; Zhang, Y.; Luo, X.-C. Biocontrol activity of recombinant aspartic protease from Trichoderma harzianum against pathogenic fungi. Enzyme Microb. Technol. 2018, 112, 35–42.
- Philip, N.V.; Koteshwara, A.; Kiran, G.A.; Raja, S.; Subrahmanyam, V.M.; Chandrashekar, H.R. Statistical optimization for coproduction of chitinase and beta 1,4-endoglucanase by chitinolytic Paenibacillus elgii PB1 having antifungal activity. Appl. Biochem. Biotechnol. 2020, 191, 135–150.
- Oyeleye, A.; Normi, Y.M. Chitinase: Diversity, limitations, and trends in engineering for suitable applications. Biosci. Rep. 2018, 38, BSR2018032300.
- Zhang, W.; Liu, Y.; Ma, J.; Yan, Q.; Jiang, Z.; Yang, S. Biochemical characterization of a bifunctional chitinase/lysozyme from Streptomyces sampsonii suitable for N-acetyl chitobiose production. Biotechnol. Lett. 2020, 42, 1489–1499.
- Li, S.; Zhang, B.; Zhu, H.; Zhu, T. Cloning and expression of the chitinase encoded by ChiKJ406136 from Streptomyces sampsonii (Millard & Burr) Waksman KJ40 and its antifungal effect. Forests 2018, 9, 699.
- Salazar, V.A.; Arranz-Trullén, J.; Navarro, S.; Blanco, J.A.; Sánchez, D.; Moussaoui, M.; Boix, E. Exploring the mechanisms of action of human secretory RNase 3 and RNase 7 against Candida albicans. Microbiol. Open 2016, 5, 830–845.
- Salazar, V.A.; Arranz-Trullén, J.; Prats-Ejarque, G.; Torrent, M.; Andreu, D.; Pulido, D.; Boix, E. Insight into the antifungal mechanism of action of human RNase N-terminus derived peptides. Int. J. Mol. Sci. 2019, 20, 4558.
- Tan, Y.; Ma, S.; Leonhard, M.; Moser, D.; Ludwig, R.; Schneider-Stickler, B. Co-immobilization of cellobiose dehydrogenase and deoxyribonuclease I on chitosan nanoparticles against fungal/bacterial polymicrobial biofilms targeting both biofilm matrix and microorganisms. Mater. Sci. Eng. C 2020, 108, 110499.
- Vidhate, R.P.; Bhide, A.J.; Gaikwad, S.M.; Giri, A.P. A potent chitin-hydrolyzing enzyme from Myrothecium verrucaria affects growth and development of Helicoverpa armigera and plant fungal pathogens. Int. J. Biol. Macromol. 2019, 141, 517–528.
- Silva, F.A.; Albuquerque, L.M.; Martins, T.F.; de Freitas, J.A.; Vasconcelos, I.M.; de Freitas, D.Q.; Moreno, F.B.M.B.; Monteiro-Moreira, A.C.O.; Oliveira, J.T.A. A peroxidase purified from cowpea roots possesses high thermal stability and displays antifungal activity against Colletotrichum gloeosporioides and Fusarium oxysporum. Biocatal. Agric. Biotechnol. 2022, 42, 102322.
- Zhang, L.; Tao, Y.; Zhao, S. A novel peroxiredoxin from the antagonistic endophytic bacterium Enterobacter sp. V1 contributes to cotton resistance against Verticillium dahliae. Plant Soil 2020, 454, 395–409.
- Pham, C.L.L.; de Francisco, B.R.; Valsecchi, I.; Dazzoni, R.; Pillé, A.; Lo, V.; Ball, S.R.; Cappai, R.; Wien, F.; Kwan, A.H.; et al. Probing structural changes during self-assembly of surface-active hydrophobin proteins that form functional amyloids in fungi. J. Mol. Biol. 2018, 430, 3784–3801.
- Valsecchi, I.; Dupres, V.; Stephen-Victor, E.; Guijarro, J.I.; Gibbons, J.; Beau, R.; Bayry, J.; Coppee, J.-Y.; Lafont, F.; Latgé, J.P.; et al. Role of hydrophobins in Aspergillus fumigatus. J. Fungi 2017, 4, 2.
- Valsecchi, I.; Lai, J.I.; Stephen-Victor, E.; Pillé, A.; Beaussart, A.; Lo, V.; Pham, C.L.L.; Aimanianda, V.; Kwan, A.H.; Duchateau, M.; et al. Assembly and disassembly of Aspergillus fumigatus conidial rodlets. Cell Surf. 2019, 5, 100023.
- Pham, C.L.L.; Rey, A.; Lo, V.; Soulès, M.; Ren, Q.; Meisl, G.; Knowles, T.P.J.; Kwan, A.H.; Sunde, M. Self-assembly of MPG1, a hydrophobin protein from the rice blast fungus that forms functional amyloid coatings, occurs by a surface-driven mechanism. Sci. Rep. 2016, 6, 25288.
- Saad, S.; Cereghetti, G.; Feng, Y.; Picotti, P.; Peter, M.; Dechant, R. Reversible protein aggregation is a protective mechanism to ensure cell cycle restart after stress. Nat. Cell Biol. 2017, 19, 1202e13.
- Beaussart, A.; Alsteens, D.; El-Kirat-Chatel, S.; Lipke, P.N.; Kucharíkova, S.; Dijck, P.V.; Dufrene, Y.F. Single-molecule imaging and functional analysis of Als adhesins and mannans during Candida albicans morphogenesis. ACS Nano 2012, 6, 10950–10964.
- Ho, V.; Herman-Bausier, P.; Shaw, C.; Conrad, K.A.; Garcia-Sherman, M.C.; Draghi, J.; Dufrene, Y.F.; Lipke, P.N.; Rauceo, J.M. An amyloid core sequence in the major Candida albicans adhesin Als1p mediates cell-cell adhesion. mBio 2019, 10, 10–128.
- Kumar, R.; Breindel, C.; Saraswat, D.; Cullen, P.J.; Edgerton, M. Candida albicans Sap6 amyloid regions function in cellular aggregation and zinc binding, and contribute to zinc acquisition. Sci. Rep. 2017, 7, 2908.
- Monniot, C.; Boisrame, A.; Costa, G.D.; Chauvel, M.; Sautour, M.; Bougnoux, M.-E.; Bellon-Fontaine, M.-N.; Dalle, F.; d’Enfert, C.; Richard, M.L. Rbt1 protein domains analysis in Candida albicans brings insights into hyphal surface modifications and Rbt1 potential role during adhesion and biofilm formation. PLoS ONE 2013, 8, e82395.
- Cabral, V.; Znaidi, S.; Walker, L.A.; Martin-Yken, H.; Dague, E.; Legrand, M.; Lee, K.; Chauvel, M.; Firon, A.; Rossignol, T.; et al. Targeted changes of the cell wall proteome influence Candida albicans ability to form single- and multi-strain biofilms. PLoS Pathog. 2014, 10, e1004542.
- Moreno-Ruiz, E.; Ortu, G.; de Groot, P.W.J.; Cottier, F.; Loussert, C.; Prevost, M.-C.; de Koster, C.; Klis, M.F.; Goyard, S.; d’Enfert, C. The GPI-modified proteins Pga59 and Pga62 of Candida albicans are required for cell wall integrity. Microbiology 2009, 155, 2004e20.
- Shanmugam, N.; Baker, M.O.; Ball, S.R.; Steain, M.; Pham, C.L.; Sunde, M. Microbial functional amyloids serve diverse purposes for structure, adhesion and defence. Biophys. Rev. 2019, 11, 287–302.
- Chernova, T.A.; Chernoff, Y.O.; Wilkinson, K.D. Yeast models for amyloids and prions: Environmental modulation and drug discovery. Molecules 2019, 24, 3388.
- Hirata, A.; Hori, Y.; Koga, Y.; Okada, J.; Sakudo, A.; Ikuta, K.; Kanaya, S.; Takano, K. Enzymatic activity of a subtilisin homolog, Tk-SP, from Thermococcus kodakarensis in detergents and its ability to degrade the abnormal prion protein. BMC Biotech. 2013, 13, 19.
- Dabbagh, F.; Negahdaripour, M.; Berenjian, A.; Behfar, A.; Mohammadi, F.; Zamani, M.; Irajie, C.; Ghasemi, Y. Nattokinase: Production and application. Appl. Microbiol. Biotechnol. 2014, 98, 9199–9206.
- Pilon, J.L.; Nash, P.B.; Arver, T.; Hoglund, D.; VerCauteren, K.C. Feasibility of infectious prion digestion using mild conditions and commercial subtilisin. J. Virol. Methods 2009, 161, 168–172.
- Saunders, S.E.; Bartz, J.C.; Vercauteren, K.C.; Bartelt-Hunt, S.L. Enzymatic digestion of chronic wasting disease prions bound to soil. Environ. Sci. Technol. 2010, 44, 4129–4135.
- McLeod, A.H.; Murdoch, H.; Dickinson, J.; Dennis, M.J.; Hall, G.A.; Buswell, C.M.; Carr, J.; Taylor, D.M.; Sutton, J.M.; Raven, N.D.H. Proteolytic inactivation of the bovine spongiform encephalopathy agent. Biochem. Biophys. Res. Commun. 2004, 317, 1165–1170.
- Dickinson, J.; Murdoch, H.; Dennis, M.J.; Hall, G.A.; Bott, R.; Crabb, W.D.; Penet, C.; Sutton, J.M.; Raven, N.D.H. Decontamination of prion protein (BSE301V) using a genetically engineered protease. J. Hosp. Infect. 2009, 72, 65–70.
- Yoshioka, M.; Miwa, T.; Horii, H.; Takata, M.; Yokoyama, T.; Nishizawa, K.; Watanabe, M.; Shinagawa, M.; Murayama, Y. Characterization of a proteolytic enzyme derived from a Bacillus strain that effectively degrades prion protein. J. Appl. Microbiol. 2007, 102, 509–515.
- Hui, Z.; Doi, H.; Kanouchi, H.; Matsuura, Y.; Mohri, S.; Nonomura, Y.; Oka, T. Alkaline serine protease produced by Streptomyces sp. degrades PrPSc. Biochem. Biophys. Res. Commun. 2004, 321, 45–50.
- Bahun, M.; Šnajder, M.; Turk, D.; Poklar Ulrih, N. Insights into the maturation of pernisine, a subtilisin-like protease from the hyperthermophilic archaeon Aeropyrum pernix. Appl. Environ. Microbiol. 2020, 86, e00971-20.
- Johnson, C.J.; Bennett, J.P.; Biro, S.M.; Duque-Velasquez, J.C.; Rodriguez, C.M.; Bessen, R.A.; Rocke, T.E. Degradation of the disease-associated prion protein by a serine protease from lichens. PLoS ONE 2011, 11, e19836.
- Chen, C.Y.; Rojanatavorn, K.; Clark, A.C.; Shih, J.C. Characterization and enzymatic degradation of Sup35NM, a yeast prion-like protein. Prot. Sci. 2005, 14, 2228–2235.
- Wang, J.J.; Borwornpinyo, R.; Odetallah, N.; Shih, J.C. Enzymatic degradation of a prion-like protein, Sup35NM-His6. Enzyme Microb. Technol. 2005, 36, 758–765.
- Sharma, R.; Gupta, R. Coupled action of γ-glutamyl transpeptidase-glutathione and keratinase effectively degrades feather keratin and surrogate prion protein, Sup 35NM. Biores. Tech. 2012, 120, 314–317.
- Rajput, R.; Gupta, R. Thermostable keratinase from Bacillus pumilus KS12: Production, chitin crosslinking and degradation of Sup35NM aggregates. Biores. Tech. 2013, 133, 118–126.
- Ningthoujam, D.S.; Mukherjee, S.; Devi, L.J.; Singh, E.S.; Tamreihao, K.; Khunjamayum, R.; Banerjee, S.; Mukhopadhyay, D. In vitro degradation of β-amyloid fibrils by microbial keratinase. Alzheimers Dement. 2019, 5, 154–163.
- Kim, N.; Lee, H.J. Redox-active metal ions and amyloid-degrading enzymes in Alzheimer’s disease. Int. J. Mol. Sci. 2021, 22, 7697.
- Manikandan, P.; Moopantakath, J.; Imchen, M.; Kumavath, R.; SenthilKumar, P.K. Identification of multi-potent protein subtilisin A from halophilic bacterium Bacillus firmus VE2. Microb. Pathog. 2021, 157, 105007.
- Kokwe, L.; Nnolim, N.E.; Ezeogu, L.I.; Sithole, B.; Nwodo, U.U. Thermoactive metallo-keratinase from Bacillus sp. NFH5: Characterization, structural elucidation, and potential application as detergent additive. Heliyon 2023, 9, e13635.
- Efremenko, E.; Aslanli, A.; Lyagin, I. Advanced situation with recombinant toxins: Diversity, production and application purposes. Int. J. Mol. Sci. 2023, 24, 4630.
- Efremenko, E.; Senko, O.; Stepanov, N.; Aslanli, A.; Maslova, O.; Lyagin, I. Quorum sensing as a trigger that improves characteristics of microbial biocatalysts. Microorganisms 2023, 11, 1395.
- Willaert, R.G. Adhesins of yeasts: Protein structure and interactions. J. Fungi 2018, 4, 119.
- Tian, X.; Ding, H.; Ke, W.; Wang, L. Quorum sensing in fungal species. Annu. Rev. Microbiol. 2021, 75, 449–469.
- Mehmood, A.; Liu, G.; Wang, X.; Meng, G.; Wang, C.; Liu, Y. Fungal quorum-sensing molecules and inhibitors with potential antifungal activity: A review. Molecules 2019, 24, 1950.
- Lee, K.; Lee, S.; Lee, S.H.; Kim, S.R.; Oh, H.S.; Park, P.K.; Choo, K.H.; Kim, Y.W.; Lee, J.K.; Lee, C.H. Fungal quorum quenching: A paradigm shift for energy savings in membrane bioreactor (MBR) for wastewater treatment. Environ. Sci. Technol. 2016, 50, 10914–10922.
- Ogawa, K.; Nakajima-Kambe, T.; Nakahara, T.; Kokufuta, E. Coimmobilization of gluconolactonase with glucose oxidase for improvement in kinetic property of enzymatically induced volume collapse in ionic gels. Biomacromolecules 2002, 3, 625–631.
- Aslanli, A.; Domnin, M.; Stepanov, N.; Efremenko, E. “Universal” antimicrobial combination of bacitracin and His6-OPH with lactonase activity, acting against various bacterial and yeast cells. Int. J. Mol. Sci. 2022, 23, 9400.
- Aslanli, A.; Domnin, M.; Stepanov, N.; Efremenko, E. Synergistic antimicrobial action of lactoferrin-derived peptides and quorum quenching enzymes. Int. J. Mol. Sci. 2023, 24, 3566.
- Hogan, D. Talking to themselves: Autoregulation and quorum sensing in fungi. Eukaryot. Cell 2006, 5, 613–619.
- Bu’LocK, J.D.; Jones, B.E.; Winskill, N. The apocarotenoid system of sex hormones and prohormones in mucorales. Pure Appl. Chem. 1976, 47, 191–202.
- Frolov, G.; Lyagin, I.; Senko, O.; Stepanov, N.; Pogorelsky, I.; Efremenko, E. Metal nanoparticles for improving bactericide functionality of usual fibers. Nanomaterials 2020, 10, 1724.
- Lyagin, I.; Stepanov, N.; Frolov, G.; Efremenko, E. Combined modification of fiber materials by enzymes and metal nanoparticles for chemical and biological protection. Int. J. Mol. Sci. 2022, 23, 1359.
- Lyagin, I.; Maslova, O.; Stepanov, N.; Presnov, D.; Efremenko, E. Assessment of composite with fibers as a support for antibacterial nanomaterials: A case study of bacterial cellulose, polylactide and usual textile. Fibers 2022, 10, 70.
- Lyagin, I.; Maslova, O.; Stepanov, N.; Efremenko, E. Degradation of mycotoxins in mixtures by combined proteinous nanobiocatalysts: In silico, in vitro and in vivo. Int. J. Biol. Macromol. 2022, 218, 866–877.
- Garces, F.; Fernández, F.J.; Montellà, C.; Penya-Soler, E.; Prohens, R.; Aguilar, J.; Baldomà, L.; Coll, M.; Badia, J.; Vega, M.C. Molecular architecture of the Mn2+-dependent lactonase UlaG reveals an RNase-like metallo-β-lactamase fold and a novel quaternary structure. J. Mol. Biol. 2010, 39, 715–729.
- González, J.M. Visualizing the superfamily of metallo-β-lactamases through sequence similarity network neighborhood connectivity analysis. Heliyon 2021, 7, e05867.
- Jang, E.-Y.; Son, Y.-J.; Park, S.-Y.; Yoo, J.-Y.; Cho, Y.-N.; Jeong, S.-Y.; Liu, S.; Son, H.-J. Improved biosynthesis of silver nanoparticles using keratinase from Stenotrophomonas maltophilia R13: Reaction optimization, structural characterization, and biomedical activity. Bioprocess Biosyst. Eng. 2018, 41, 381–393.
- Abo-Zaid, G.; Abdelkhalek, A.; Matar, S.; Darwish, M.; Abdel-Gayed, M. Application of bio-friendly formulations of chitinase-producing Streptomyces cellulosae Actino 48 for controlling peanut soil-borne diseases caused by Sclerotium rolfsii. J. Fungi 2021, 7, e167.
- Gaonkar, S.K.; Furtado, I.J. Biorefinery-fermentation of agro-wastes by Haloferax lucentensis GUBF-2 MG076878 to haloextremozymes for use as biofertilizer and biosynthesizer of AgNPs. Waste Biomass Valorization 2022, 13, 1117–1133.
- Guilger-Casagrande, M.; Germano-Costa, T.; Pasquoto-Stigliani, T.; Fraceto, L.F.; de Lima, R. Biosynthesis of silver nanoparticles employing Trichoderma harzianum with enzymatic stimulation for the control of Sclerotinia sclerotiorum. Sci. Rep. 2019, 9, e14351.
- Guilger-Casagrande, M.; Germano-Costa, T.; Bilesky-José, N.; Pasquoto-Stigliani, T.; Carvalho, L.; Fraceto, L.F.; de Lima, R. Influence of the capping of biogenic silver nanoparticles on their toxicity and mechanism of action towards Sclerotinia sclerotiorum. J. Nanobiotechnol. 2021, 19, e53.
- Dror, Y.; Ophir, C.; Freeman, A. Silver-enzyme hybrids as wide-spectrum antimicrobial agents. In Innovations and Merging Technologies in Wound Care; Gefen, A., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 293–307.
- Xia, S.; Xu, Y.; Hoy, R.; Zhang, J.; Qin, L.; Li, X. The notorious soilborne pathogenic fungus Sclerotinia sclerotiorum: An update on genes studied with mutant analysis. Pathogens 2020, 9, e27.
- Chet, I.; Henis, Y. Effect of catechol and disodium EDTA on melanin content of hyphal and sclerotial walls of Sclerotium rolfsii sacc. and the role of melanin in the susceptibility of these walls to β-(1→3) glucanase and chitinase. Soil Biol. Biochem. 1969, 1, 131–138.
- Melo, B.S.; Voltan, A.R.; Arruda, W.; Lopes, F.A.C.; Georg, R.C.; Ulhoa, C.J. Morphological and molecular aspects of sclerotial development in the phytopathogenic fungus Sclerotinia sclerotiorum. Microbiol. Res. 2019, 229, e126326.
- Yu, D.; Wang, Y.; Zhang, J.; Yu, Q.; Liu, S.; Li, M. Synthesis of the ternary nanocomposites composed of zinc 2-methylimidazolate frameworks, lactoferrin and melittin for antifungal therapy. J. Mater. Sci. 2022, 57, 16809–16819.
- Babina, S.E.; Kanyshkova, T.G.; Buneva, V.N.; Nevinsky, G.A. Lactoferrin is the major deoxyribonuclease of human milk. Biochemistry 2004, 69, 1006–1015.
- Fernandes, K.E.; Weeks, K.; Carter, D.A. Lactoferrin is broadly active against yeasts and highly synergistic with amphotericin B. Antimicrob. Agents Chemother. 2020, 64, e02284-19.




