
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Italia Di Liegro | -- | 1819 | 2023-07-20 11:19:14 | | | |
| 2 | Alfred Zheng | Meta information modification | 1819 | 2023-07-21 03:51:00 | | |
Video Upload Options
All the cells of an organism contain the same genome. However, each cell expresses only a minor fraction of its potential and, in particular, the genes encoding the proteins necessary for basal metabolism and the proteins responsible for its specific phenotype. The ability to use only the right and necessary genes involved in specific functions depends on the structural organization of the nuclear chromatin, which in turn depends on the epigenetic history of each cell, which is stored in the form of a collection of DNA and protein modifications. Among these modifications, DNA methylation and many kinds of post-translational modifications of histones play a key role in organizing the complex indexing of usable genes.
1. Introduction
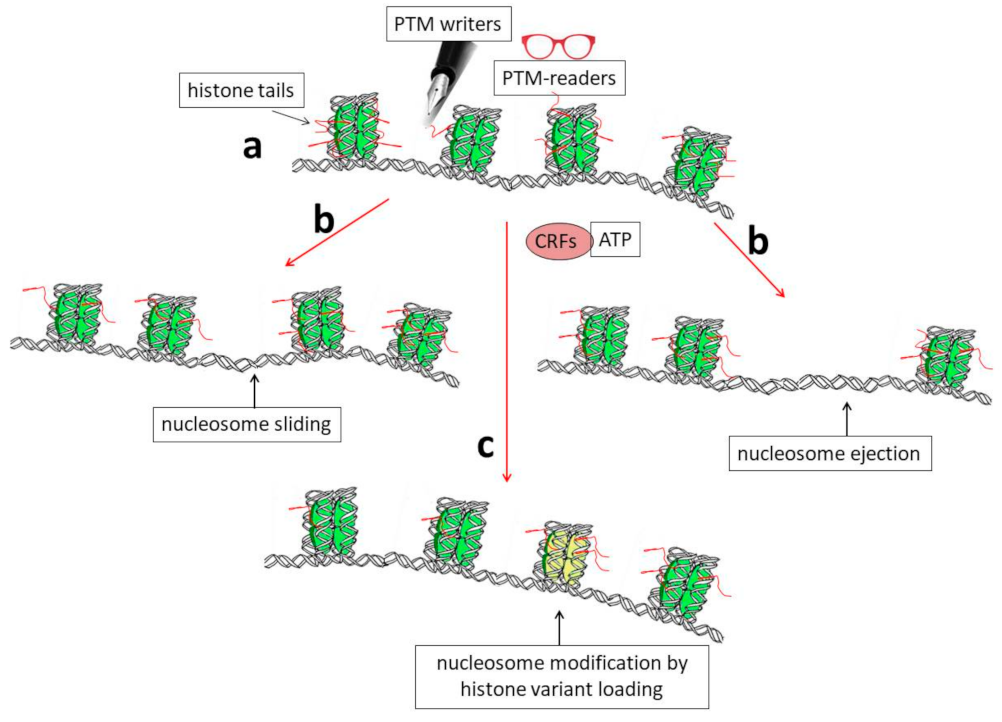
2. General Properties of Genes Encoding Histone Variants
References
- Gurdon, J.B.; Elsdale, T.R.; Fischberg, M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 1958, 182, 64–65.
- Kornberg, R.D. Chromatin structure: A repeating unit of histones and DNA. Science 1974, 184, 868–871.
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260.
- Felsenfeld, G. A brief history of epigenetics. Cold Spring Harb. Perspect. Biol. 2014, 6, a018200.
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500.
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220.
- Sokolova, V.; Sarkar, S.; Tan, D. Histone variants and chromatin structure, update of advances. Comput. Struct. Biotechnol. J. 2022, 21, 299–311.
- Hergeth, S.P.; Schneider, R. The H1 linker histones: Multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015, 16, 1439–1453.
- Waddington, C.H. The epigenotype. 1942. Int. J. Epidemiol. 2012, 41, 10–13.
- Kim, U.; Lee, D.-S. Epigenetic Regulations in Mammalian Cells: Roles and Profiling Techniques. Mol. Cells 2023, 46, 86–98.
- Kato, S.; Yokoyama, A.; Fujiki, R. Nuclear receptor coregulators merge transcriptional coregulation with epigenetic regulation. Trends Biochem. Sci. 2011, 36, 272–281.
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45.
- Zhu, H.; Wang, G.; Qian, J. Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 2016, 17, 551–565.
- Luo, C.; Hajkova, P.; Ecker, J.R. Dynamic DNA methylation: In the right place at the right time. Science 2018, 361, 1336–1340.
- Jiang, D.; Li, T.; Guo, C.; Tang, T.S.; Liu, H. Small molecule modulators of chromatin remodeling: From neurodevelopment to neurodegeneration. Cell Biosci. 2023, 13, 10.
- Shindo, Y.; Brown, M.G.; Amodeo, A.A. Versatile roles for histones in early development. Curr. Opin. Cell Biol. 2022, 75, 102069.
- Peng, J.; Zhang, W.J.; Zhang, Q.; Su, Y.H.; Tang, L.P. The dynamics of chromatin states mediated by epigenetic modifications during somatic cell reprogramming. Front. Cell. Dev. Biol. 2023, 11, 1097780.
- Simon, L.; Probst, A.V. Maintenance and dynamic reprogramming of chromatin organization during development. Plant J. 2023.
- Marzluff, W.F.; Gongidi, P.; Woods, K.R.; Jin, J.; Maltais, L.J. The human and mouse replication-dependent histone genes. Genomics 2002, 80, 487–498.
- Marzluff, W.F.; Wagner, E.J.; Duronio, R.J. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat. Rev. Genet. 2008, 9, 843–854.
- Amatori, S.; Tavolaro, S.; Gambardella, S.; Fanelli, M. The dark side of histones: Genomic organization and role of oncohistones in cancer. Clin. Epigenetics 2021, 13, 71.
- Talbert, P.B.; Henikoff, S. Histone variants at a glance. J. Cell Sci. 2021, 134, jcs244749.
- Castiglia, D.; Cestelli, A.; Scaturro, M.; Nastasi, T.; Di Liegro, I. H1.0 and H3.3B mRNA levels in developing rat brain. Neurochem. Res. 1994, 19, 1531–1537.
- Scaturro, M.; Cestelli, A.; Castiglia, D.; Nastasi, T.; Di Liegro, I. Posttranscriptional Regulation of H1.0 and H3.3B histone genes in differentiating rat cortical neurons. Neurochem. Res. 1995, 20, 969–976.
- Castiglia, D.; Scaturro, M.; Nastasi, T.; Cestelli, A.; Di Liegro, I. PIPPin, a putative RNA-binding protein specifically expressed in the rat brain. Biochem. Biophys. Res. Commun. 1996, 218, 390–394.
- Scaturro, M.; Nastasi, T.; Raimondi, L.; Bellafiore, M.; Cestelli, A.; Di Liegro, I. H1(0) RNA-binding proteins specifically expressed in the rat brain. J. Biol. Chem. 1998, 273, 22788–22791.
- Nastasi, T.; Scaturro, M.; Bellafiore, M.; Raimondi, L.; Beccari, S.; Cestelli, A.; Di Liegro, I. PIPPin is a brain-specific protein that contains a cold-shock domain and binds specifically to H1 degrees and H3.3 mRNAs. J. Biol Chem. 1999, 274, 24087–24093.
- Sala, A.; Scaturro, M.; Proia, P.; Schiera, G.; Balistreri, E.; Aflalo-Rattenbach, R.; Créau, N.; Di Liegro, I. Cloning of a rat-specific long PCP4/PEP19 isoform. Int. J. Mol. Med. 2007, 19, 501–509.
- Saladino, P.; Di Liegro, C.M.; Proia, P.; Sala, A.; Schiera, G.; Lo Cicero, A.; Di Liegro, I. RNA-binding activity of the rat calmodulin-binding PEP-19 protein and of the long PEP-19 isoform. Int. J. Mol. Med. 2012, 29, 141–145.
- Weaver, K.J.; Holt, R.A.; Henry, E.; Pletcher, S.D. Effects of hunger on neuronal histone modifications slow aging in Drosophila. Science 2023, 380, 625–632.
- Kumar, V.C.; Pai, R. Genes of the month: H3.3 histone genes: H3F3A and H3F3B. J. Clin. Pathol. 2021, 74, 753–758.
- Bryant, L.; Sangree, A.; Clark, K.; Bhoj, E. Histone 3.3-related chromatinopathy: Missense variants throughout H3-3A and H3-3B cause a range of functional consequences across species. Hum. Genet. 2023.
- Bush, K.; Cervantes, V.; Yee, J.Q.; Klein, R.H.; Knoepfler, P.S. A knockout-first model of H3f3a gene targeting leads to developmental lethality. Genesis 2023, 61, e23507.
- Bachu, M.; Tamura, T.; Chen, C.; Narain, A.; Nehru, V.; Sarai, N.; Ghosh, S.B.; Ghosh, A.; Kavarthapu, R.; Dufau, M.L.; et al. A versatile mouse model of epitope-tagged histone H3.3 to study epigenome dynamics. J. Biol. Chem. 2019, 294, 1904–1914.
- Elsaesser, S.J.; Goldberg, A.D.; Allis, C.D. New functions for an old variant: No substitute for histone H3.3. Curr. Opin. Genet. Dev. 2010, 20, 110–117.
- Filipescu, D.; Szenker, E.; Almouzni, G. Developmental roles of histone H3 variants and their chaperones. Trends Genet. 2013, 29, 630–640.
- Delaney, K.; Almouzni, G. Transcription-coupled H3.3 recycling: A link with chromatin states. Semin. Cell Dev. Biol. 2023, 135, 13–23.
- Lewis, P.W.; Elsaesser, S.J.; Noh, K.M.; Stadler, S.C.; Allis, C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl Acad. Sci. USA 2010, 107, 14075–14080.
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010, 140, 678–691.
- Smith, R.; Susor, A.; Ming, H.; Tait, J.; Conti, M.; Jiang, Z.; Lin, C.-J. The H3.3 chaperone Hira complex orchestrates oocyte developmental competence. Development 2022, 149, dev200044.
- Yang, Y.; Zhang, L.; Xiong, C.; Chen, J.; Wang, L.; Wen, Z.; Yu, J.; Chen, P.; Xu, Y.; Jin, J.; et al. HIRA complex presets transcriptional potential through coordinating depositions of the histone variants H3.3 and H2A.Z on the poised genes in mESCs. Nucleic Acids Res. 2022, 50, 191–206.
- Truch, J.; Downes, D.J.; Scott, C.; Gür, E.R.; Telenius, J.M.; Repapi, E.; Schwessinger, R.; Gosden, M.; Brown, J.M.; Taylor, S.; et al. The chromatin remodeller ATRX facilitates diverse nuclear processes, in a stochastic manner, in both heterochromatin and euchromatin. Nat. Commun. 2022, 13, 3485.
- Ahmad, K.; Henikoff, S. The histone Variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 2002, 9, 1191–1200.
- Chen, P.; Zhao, J.; Wang, Y.; Wang, M.; Long, H.; Liang, D.; Huang, L.; Wen, Z.; Li, W.; Li, X.; et al. H3.3 actively marks enhancers and primes gene transcription via opening higher ordered chromatin. Genes Dev. 2013, 27, 2109–2124.
- Deaton, A.M.; Gomez-Rodriguez, M.; Mieczkowski, J.; Tolstorukov, M.Y.; Kundu, S.; Sadreyev, R.I.; Jansen, L.E.; Kingston, R.E. Enhancer regions show high histone H3.3 turnover that changes during differentiation. Elife 2016, 5, e15316.
- Shi, L.; Wen, H.; Shi, X. The histone variant H3.3 in transcriptional regulation and human disease. J. Mol. Biol. 2017, 429, 1934–1945.
- Reske, J.J.; Wilson, M.R.; Armistead, B.; Harkins, S.; Perez, C.; Hrit, J.; Adams, M.; Rothbart, S.B.; Missmer, S.A.; Fazleabas, A.T.; et al. ARID1A-dependent maintenance of H3.3 is required for repressive CHD4-ZMYND8 chromatin interactions at super-enhancers. BMC Biol. 2022, 20, 209.
- Wong, L.H.; Ren, H.; Williams, E.; McGhie, J.; Ahn, S.; Sim, M.; Tam, A.; Earle, E.; Anderson, M.A.; Mann, J.; et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009, 19, 404–414.
- Udugama, M.; Chang, F.T.M.; Chan, F.L.; Tang, M.C.; Pickett, H.A.; McGhie, J.D.R.; Mayne, L.; Collas, P.; Mann, J.R.; Wong, L.H. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 2015, 43, 10227–10237.
- Fang, L.; Chen, D.; Zhang, J.; Li, H.; Bradford, B.; Jin, C. Potential functions of histone H3.3 lysine 56 acetylation in mammals. Epigenetics 2022, 17, 498–517.
- Schoberleitner, I.; Mertens, B.; Bauer, I.; Lusser, A. Regulation of sensory perception and motor abilities by brain-specific action of chromatin remodelling factor CHD1. Front. Mol. Neurosci. 2022, 15, 840966.
- Sakai, A.; Schwartz, B.E.; Goldstein, S.; Ahmad, K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr. Biol. 2009, 19, 1816–1820.
- Armache, A.; Yang, S.; Martínez de Paz, A.; Robbins, L.E.; Durmaz, C.; Cheong, J.Q.; Ravishankar, A.; Daman, A.W.; Ahimovic, D.J.; Klevorn, T.; et al. Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature 2020, 583, 852–857.
- Chen, J.; Horton, J.; Sagum, C.; Zhou, J.; Cheng, X.; Bedford, M.T. Histone H3 N-terminal mimicry drives a novel network of methyl-effector interactions. Biochem. J. 2021, 478, 1943–1958.
- Udugama, M.; Vinod, B.; Chan, F.L.; Hii, L.; Garvie, A.; Collas, P.; Kalitsis, P.; Steer, D.; Das, P.P.; Tripathi, P.; et al. Histone H3.3 phosphorylation promotes heterochromatin formation by inhibiting H3K9/K36 histone demethylase. Nucleic Acids Res. 2022, 50, 4500–4514.
- Martire, S.; Gogate, A.A.; Whitmill, A.; Tafessu, A.; Nguyen, J.; Teng, Y.C.; Tastemel, M.; Banaszynski, L.A. Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat. Genet. 2019, 51, 941–946.
- Tafessu, A.; O’Hara, R.; Martire, S.; Dube, A.L.; Saha, P.; Gant, V.U.; Banaszynski, L.A. H3.3 contributes to chromatin accessibility and transcription factor binding at promoter-proximal regulatory elements in embryonic stem cells. Genome Biol. 2023, 24, 25.





