Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Amol Tandon | -- | 3669 | 2023-07-19 13:37:16 | | | |
| 2 | Camila Xu | Meta information modification | 3669 | 2023-07-20 03:09:23 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tandon, A.; Kuriappan, J.A.; Dubey, V. MYC Deregulation in Burkitt Lymphoma. Encyclopedia. Available online: https://encyclopedia.pub/entry/46979 (accessed on 05 March 2026).
Tandon A, Kuriappan JA, Dubey V. MYC Deregulation in Burkitt Lymphoma. Encyclopedia. Available at: https://encyclopedia.pub/entry/46979. Accessed March 05, 2026.
Tandon, Amol, Jissy Akkarapattiakal Kuriappan, Vaibhav Dubey. "MYC Deregulation in Burkitt Lymphoma" Encyclopedia, https://encyclopedia.pub/entry/46979 (accessed March 05, 2026).
Tandon, A., Kuriappan, J.A., & Dubey, V. (2023, July 19). MYC Deregulation in Burkitt Lymphoma. In Encyclopedia. https://encyclopedia.pub/entry/46979
Tandon, Amol, et al. "MYC Deregulation in Burkitt Lymphoma." Encyclopedia. Web. 19 July, 2023.
Copy Citation
MYC deregulation, a cardinal event in Burkitt lymphoma (BL) pathogenesis, necessitates the elucidation of the molecular mechanisms governing MYC activation to devise innovative and effective therapeutic strategies.
MYC
MYC translocation
MYC deregulation
Burkitt lymphoma
1. Introduction
Burkitt lymphoma (BL) is a highly aggressive and rapidly advancing subtype of non-Hodgkin’s lymphoma, hallmarked by elevated proliferation rates and substantial genetic instability. Burkitt lymphoma (BL) has several notable distinctions in cancer research. It was the first human tumor found to be linked with a virus [1] and one of the earliest tumors identified to have a chromosomal translocation activating an oncogene [2]. Additionally, it was the first lymphoma reported in association with HIV infection [3]. Moreover, it was the first pediatric tumor to show a response to chemotherapy alone [4]. In regions with endemic malaria, such as equatorial Africa, Brazil, and Papua New Guinea, BL is the most prevalent childhood cancer [5][6].
A defining feature of BL is the presence of chromosomal translocations involving the MYC gene, which encodes a pivotal transcription factor that orchestrates cell growth, differentiation, and apoptosis [7][8]. The MYC oncogene significantly contributes to many human cancers by regulating myriad genes. Its influence extends beyond cellular biology to impact host immunity and the tumor microenvironment [9][10]. It is activated through various methods—genetic, epigenetic, and post-translational—leading to altered expression, and thus influences different types of cancer [11]. Common genomic changes, such as gene amplification, chromosomal translocations, and mutations, can escalate MYC expression [12][13]. The gene further influences cancer cells by managing processes, such as growth, differentiation, and metabolism [14][15][16]. Although MYC expression is typically restrained in healthy cells, its heightened levels in cancer cells can trigger apoptosis. Nevertheless, its activation bypasses these physiological checks, thereby promoting cancer via several interconnected mechanisms [9][17][18]. The regulation of MYC and the diverse range of its targets is depicted in Figure 1. MYC overexpression in Burkitt lymphoma is commonly facilitated by chromosomal translocations, such as the MYC-immunoglobulin heavy chain (IgH) [19][20]. This translocation culminates in unrestrained MYC activation, fostering cell proliferation and impeding apoptosis through intricate and diverse mechanisms that entail interactions with a plethora of proteins and regulatory factors [21].
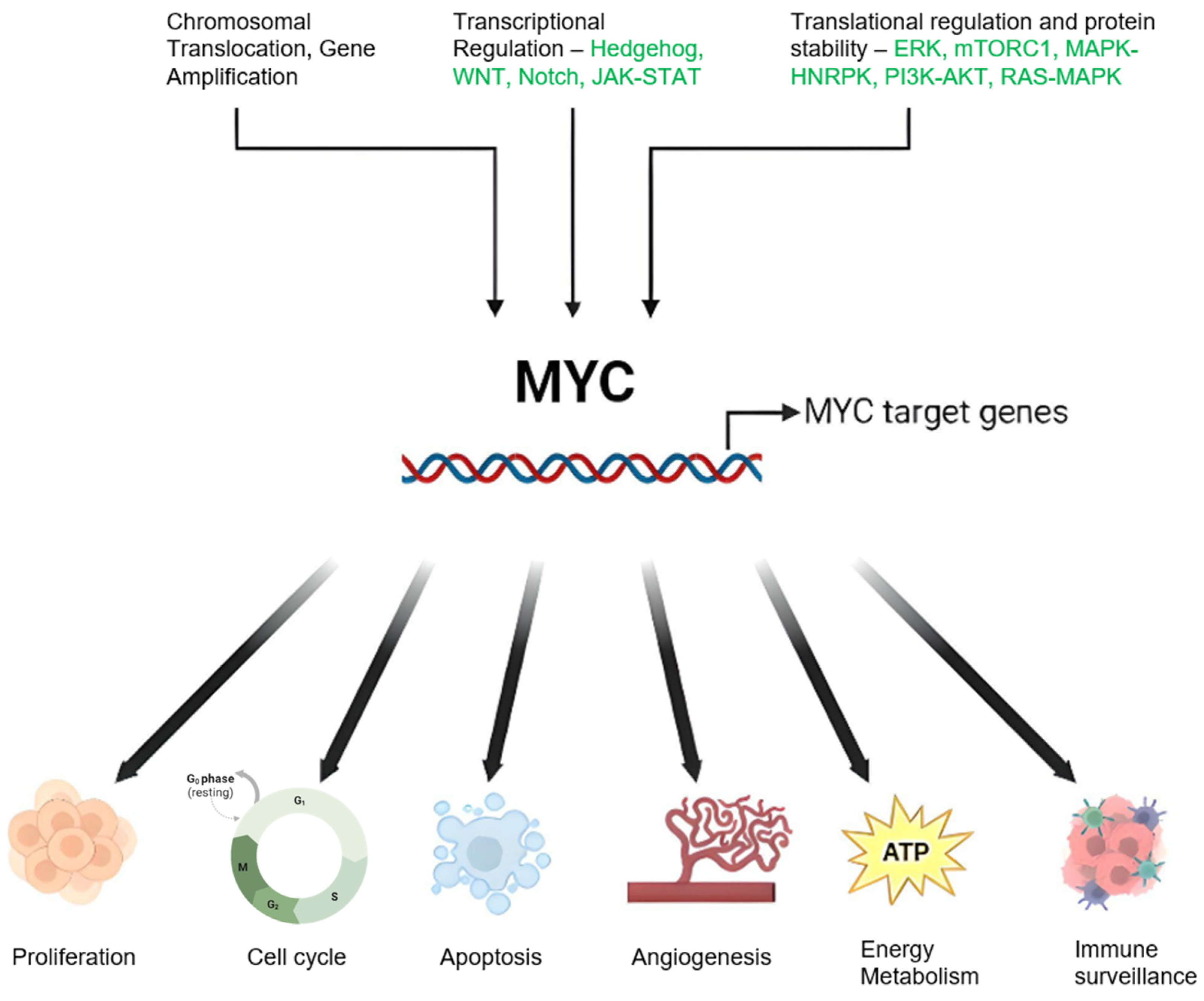
Figure 1. Schematic Representation of MYC Regulation and Function in Cancer: Genetic alterations including chromosomal translocations and genomic amplifications result in elevated MYC mRNA expression. Modifications in upstream regulatory pathways can also influence MYC oncogene transcription. MYC protein stability is enhanced through post-translational modifications, notably preferential phosphorylation at serine 62 (S62) over threonine 58 (T58), inhibiting degradation and stimulating the MYC pathway. MYC orchestrates a variety of cancer cell-intrinsic and host-dependent pathways to foster cancer cell growth and survival, with key processes including proliferation, metabolism, protein, and ribosomal biosynthesis. Conversely, MYC suppresses cellular protective mechanisms, such as immune surveillance, and thus aids in cancer progression. Notably, MYC can paradoxically trigger cellular processes, such as apoptosis that potentially threaten cancer cell survival. The ultimate cell fate is determined by the intricate equilibrium between these events and the specific cellular context. Created with BioRender.com, accessed on 20 May 2023.
In addition to chromosomal translocations, MYC point mutations have been identified in BL. Present in approximately 30% of BL cases [22], these mutations lead to increased MYC protein stability and activity, correlating with poor prognosis. Given the central role MYC occupies in BL development and progression, there is a burgeoning interest in devising MYC-targeted therapies. However, due to MYC’s transcription factor nature and the absence of a well-defined active region, the development of small compounds targeting MYC presents a formidable challenge [8].
2. MYC Deregulation in Burkitt Lymphoma: Mechanisms and Implications
- (1)
-
Chromosomal Translocations: In Burkitt lymphoma (BL), the most common mechanism of MYC deregulation involves chromosomal translocations that place the MYC gene under the control of immunoglobulin (Ig) enhancer elements, leading to constitutive activation of MYC expression in B-cells [23][24][25][26]. The translocation t(8;14)(q24;q32), found in approximately 80% of BL cases, is the most frequent MYC translocation in this disease [27][28]. Additional MYC translocations in BL include t(2;8)(p12;q24) and t(8;22)(q24;q11) [28][29][30]. Constitutive MYC overexpression drives uncontrolled cell proliferation, a defining feature of BL.
- (2)
-
Point Mutations: In addition to chromosomal translocations, point mutations in MYC have been identified in BL. The T58A mutation increases MYC protein stability by inhibiting its degradation via the ubiquitin–proteasome pathway and enhances MYC transcriptional activity by increasing its association with transcriptional co-activators, such as TRRAP and p300/CBP [31][32].
- (3)
-
Genomic Instability: MYC translocation also contributes to the genomic instability of BL cells. MYC-induced DNA replication stress can cause DNA double-strand breaks, leading to chromosomal rearrangements and mutations [33][34]. This instability can contribute to the clonal evolution of BL and the acquisition of additional mutations that drive tumor progression and resistance to therapy [35].
- (4)
-
Signaling Pathways: The molecular mechanisms of MYC-induced transformation in BL are complex and involve the deregulation of multiple signaling pathways. MYC regulates the expression of genes involved in several growth/proliferation regulating signaling pathways, including the Wnt/β-catenin, NF-κB, and PI3K/Akt/mTOR pathways [36][37]. Deregulation of these pathways can contribute to the aggressive behavior of BL and resistance to therapy.
- (5)
-
MYC and Cancer Metabolism: MYC has been implicated in cancer metabolism, particularly the Warburg effect, where it promotes glucose metabolism and aerobic glycolysis in cancer cells [38][39]. This metabolic adaptation enables cancer cells to withstand nutrient scarcity and hypoxic environments. MYC also plays a role in mitochondrial biogenesis and regulation in response to growth signals and cell cycle progression [40][41][42]. This provides an opportunity for therapeutic targeting of mitochondrial factors regulated by MYC in the context of the Warburg effect in cancer.
- (6)
-
MYC and Immune Regulation: Studies on human patient samples and transgenic mouse models have provided evidence that MYC is involved in the regulation of innate immune regulator CD-47 and the well-known adaptive immune checkpoint PD-L1 [43][44][45]. Saravia et. al [46] found that MYC also promotes the differentiation and activation of regulatory T-cells, which suppress immune responses and promote tumor growth. MYC influences the expression of genes involved in T-cell activation while suppressing the expression of genes involved in T-cell differentiation and function [46].
Figure 2 depicts data obtained from cBio Cancer Genomics Portal, where MYC altered lymphomas are shown to have a significantly worse prognosis than patients with wild-type MYC. Since MYC translocations are found in nearly all Burkitt lymphoma cases, it is difficult to establish a direct correlation between MYC alterations and survival outcomes. Instead, other factors, such as disease stage at diagnosis, patient age, performance status, lactate dehydrogenase levels, and response to treatment, are more commonly used to assess prognosis and survival in Burkitt lymphoma. Table 1 lists the various mechanisms by which MYC could be deregulated followed by detailed discussions on these topics.
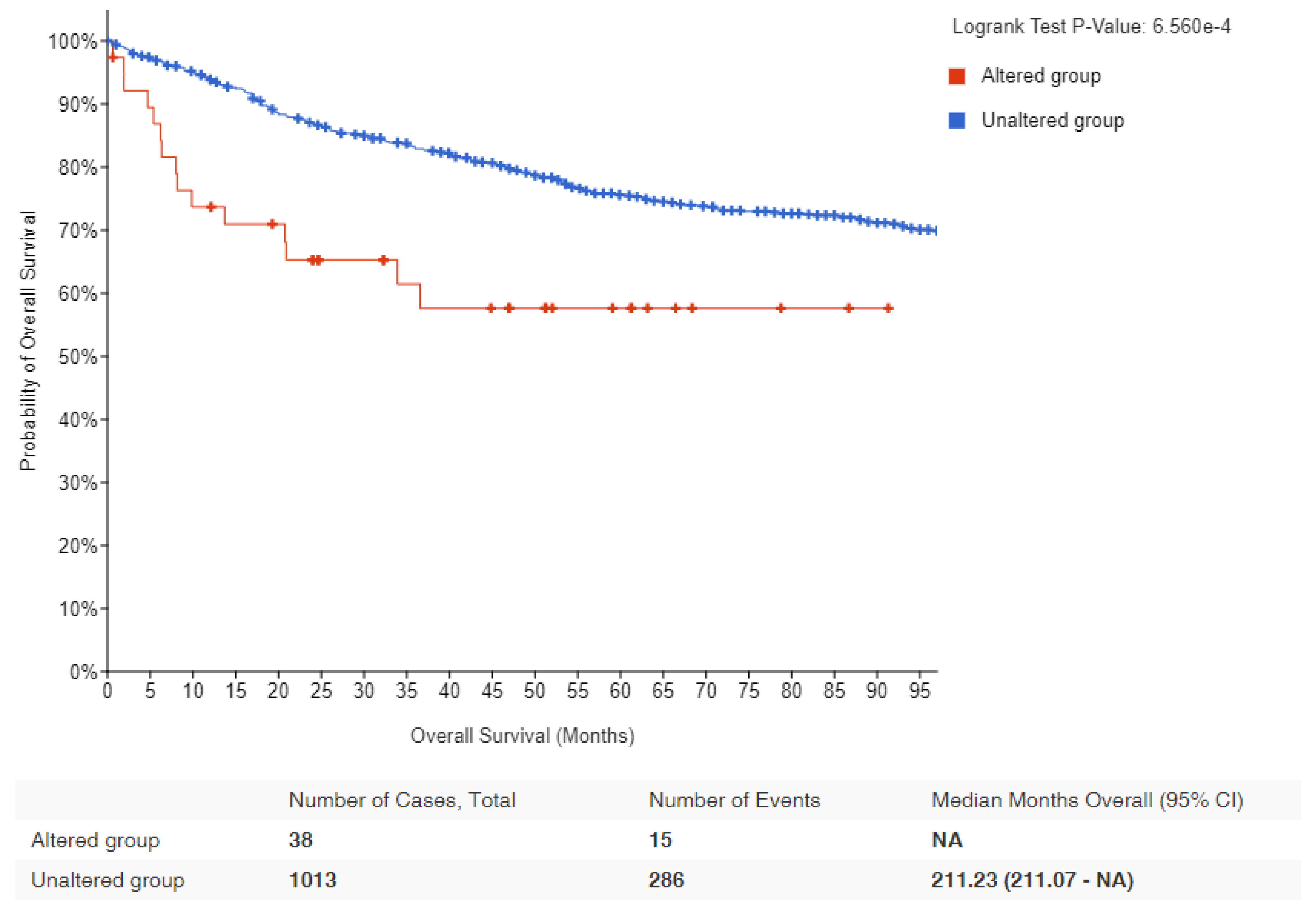
Figure 2. Kaplan–Meier survival estimates in lymphomas with MYC alteration: Data obtained from cBioPortal for lymphoma patients showing the probability of overall survival in MYC altered (red) and unaltered (blue) groups. Logrank p-value and number of samples analyzed for each group are indicated [47][48].
Table 1. Summary of various mechanisms by which MYC could be deregulated.
| Mechanism | Description |
|---|---|
| Gene Amplification | This is when there is an increase in the number of copies of the MYC gene, leading to overproduction of MYC protein. |
| Chromosomal Translocation |
This is when the MYC gene is moved to a different chromosome, often leading to its inappropriate activation. In Burkitt lymphoma, for example, MYC is often translocated to the immunoglobulin heavy or light chain loci, which are highly active regions of the genome. |
| Mutation | Mutations in the MYC gene or in the genes that regulate MYC can lead to increased MYC activity. |
| Deregulation of Transcriptional Control |
Disruptions in the normal mechanisms that control the transcription of the MYC gene can lead to overproduction of the MYC protein. |
| Post-transcriptional and Post-translational Modifications |
Changes that occur to MYC mRNA or MYC protein after transcription and translation, respectively, can increase the stability, abundance, or activity of MYC. |
| Deregulation of MYC Degradation |
Normally, MYC protein is rapidly degraded to maintain proper control of its levels. Disruptions in these degradation pathways can lead to increased levels of MYC. |
2.1. Mechanism of MYC Translocation and Its Significance in Accumulating Mutations on the Translocated Allele
2.1.1. Mechanism of IgH-MYC Translocation
Translocation occurs when chromosomes break and reattach to different chromosomes. In Burkitt lymphoma, one end of chromosome 8, where the MYC gene resides, translocates to chromosome 14, where the IgH gene is located. This leads to the fusion of the two genes. Normally, the MYC gene regulates cell division, but when it is fused with the IgH gene, it is then placed under the control of IgH’s regulatory elements. This results in overexpression of the MYC gene, leading to uncontrolled cell growth and proliferation, a key feature of lymphoma [11][49].
This translocation is often linked to errors in the somatic hypermutation process, a mechanism where B-cells modify their antibody genes to produce high-affinity antibodies. Normally, this process is tightly regulated, but errors can lead to off-target mutations, including the IgH-MYC translocation.
In the context of chromosomal rearrangements leading to the translocation of proto-oncogenes into transcription-active regions, the study by Stasevich et al. [50] highlighted the role of non-coding enhancer RNAs (eRNAs) as regulatory mechanisms affecting the oncogenes upon translocation [50]. This study emphasizes the role of non-coding enhancer RNAs (eRNAs) in regulating the activity of oncogenes following translocation. The researchers sought to identify eRNAs that might influence MYC transcription during IgH-MYC translocation in Burkitt lymphoma, with an emphasis on eRNAs that are potentially oncogenic, located at the IgH locus, and predominantly expressed in B-cells. The results from this study demonstrated that eRNA AL928768.3 is a critical player in Burkitt lymphoma development. Importantly, the study’s findings also suggest that AL928768.3, along with other yet-to-be-discovered eRNAs, could potentially serve as tissue-specific targets for cancer therapeutics [50]. This opens up a promising new avenue for the development of targeted treatments for lymphomas, particularly Burkitt lymphoma.
2.1.2. Risk Factors for Burkitt Lymphoma and IgH-MYC Translocation
Risk factors for Burkitt lymphoma, and thus the IgH-MYC translocation, include:
-
Infection with the Epstein–Barr virus (EBV): Most people are infected with EBV at some point in their lives, but in rare cases, it can increase the risk of Burkitt lymphoma. It is believed that the virus may contribute to the occurrence of the IgH-MYC translocation [51].
-
Malaria: In endemic regions (such as sub-Saharan Africa), chronic malaria infection weakens the immune system and is thought to contribute to the higher incidence of Burkitt lymphoma [52].
-
Immune suppression: Individuals with weakened immune systems (such as those with HIV/AIDS or organ transplant recipients) have an increased risk [53].
2.1.3. Mutations Accumulate in the Translocated MYC Allele
The translocation of the c-MYC gene into the immunoglobulin heavy chain (IgH) locus in Burkitt lymphoma (BL) has been reported to result in a high number of mutations due to its exposure to the antibody hypermutation mechanism [54][55][56][57][58]. Bemark and Neuberger [54] demonstrated that the translocated c-MYC undergoes constitutive hypermutation in the Ramos BL cell line at a rate comparable to that of immunoglobulin variable (IgV) genes. This study found that the non-random distribution of mutations stems from the intrinsic bias of the mutational process, rather than a skewing effect of selection, and provided insights into the cis-acting sequences required for mutability. Specifically, IgH sequences downstream of the Sμ region were sufficient, and the IgH intronic enhancer was not necessary.
Nearly two decades later, ref. [59] examined the effect of Phorbol 12-myristate 13-acetate (PMA) stimulation on the expression of the MYC gene in Ramos BL wild-type (WT) and ADP-dependent glucokinase (ADPGK) knock-out (KO) cells. The researchers discovered a larger accumulation of mutations in MYC transcripts in Ramos WT cells compared to ADPGK KO cells upon PMA stimulation. These mutations displayed a preference for AGC triplets, indicating the influence of immunoglobulin hypermutation and an organized mutational machinery. Interestingly, sequencing of stimulated Ramos BL cells revealed the recovery of the wild-type MYC sequence in nearly half of the cases, which contrasts the findings of [54]. Additionally, MYC expression increased during activation and decreased during differentiation as B-cells transitioned from an activated state to a plasma-cell-like differentiation phase, suggesting a functional role for MYC in providing energy stimulation to differentiating B-cells.
Tandon et al. [59] also found that PMA stimulation increased MYC transcript levels in Ramos cells, but ADPGK KO cells showed a smaller elevation in MYC expression compared to WT cells. The study revealed that the accumulation of mutations is dependent on cell transcriptional activity rather than activation-induced cytidine deaminase (AID) expression levels. This finding points to a role for MYC in offering an evolutionary growth advantage to cancer cells in vitro, characterized by a higher rate of proliferation and increased transcriptional activity at the translocated MYC locus.
In a separate study, ref. [60] investigated the mutational landscape of BL using high-throughput sequencing. They identified recurrent mutations in genes associated with the B-cell receptor signaling pathway, chromatin remodeling, and DNA damage response. This research not only provided valuable insights into the genetic mechanisms underlying MYC-driven lymphomagenesis in BL, but also highlighted potential therapeutic targets for future treatment strategies.
2.2. Additional Genetic Alterations in Burkitt Lymphoma
Although MYC deregulation is a hallmark of Burkitt lymphoma (BL), it alone is insufficient to cause the disease. Additional genetic modifications are necessary for the transformation of B-cells into malignant lymphomas [61]. These additional mutations are generally thought to occur independently of the IgH-MYC translocation, namely, the IgH-MYC translocation does not directly cause these additional mutations to occur. However, it is worth noting that the overexpression of MYC might indirectly contribute to a higher mutation rate. MYC overexpression can lead to increased cell proliferation and DNA replication, which can increase the chance of errors occurring during DNA replication. Furthermore, MYC can induce a state of “replicative stress”, which can lead to DNA damage and genomic instability, further increasing the potential for additional mutations.
2.2.1. TP53 Mutations and Its Prognostic Importance in BL
TP53 is a tumor suppressor gene that encodes a transcription factor that regulates cell cycle arrest, apoptosis, and DNA repair in response to cellular stress. TP53 is frequently altered in various cancers, including BL, where it is one of the most common and important co-alterations, but its role and impact on BL biology and clinical outcome are not fully understood [62][63].
Several studies have investigated the frequency, type, and prognostic value of TP53 abnormalities (mutations, deletions, and/or copy number neutral loss of heterozygosity) in BL, using different methods, such as sequencing, fluorescence in situ hybridization (FISH), immunohistochemistry, and gene expression analysis [62][63][64]. The reported prevalence of TP53 abnormalities in BL ranges from 20% to 70%, depending on the cohort, subtype, and detection method [62][63][64]. TP53 abnormalities are more common in adult than in pediatric BL, and in immunodeficiency-associated than in endemic or sporadic BL [28] TP53 abnormalities are also more frequent in cases with high-risk features, such as high lactate dehydrogenase (LDH), bone marrow or central nervous system involvement, or double-hit or triple-hit status (concurrent rearrangements of MYC, BCL2, and/or BCL6) [62].
In a comprehensive study conducted by Burkhardt et al. [65], they analyzed a large cohort of pediatric and adult Burkitt lymphoma (BL) cases. They discovered that the mutational profile of these cancers varies by age group. A critical finding was the significant prognostic impact of recurrent mutations in the TP53 gene, which are associated with diminished expression of TP53wt protein in lymphoma samples. This lack of TP53wt expression and presence of TP53 mutations were linked to decreased survival rates, suggesting the central role of TP53 in the disease’s progression. The study further implies that new therapeutic strategies could be developed to target this dysfunctional TP53, potentially by inhibiting its upstream regulators, MDM2 and MDM4. This could decrease the ubiquitination and subsequent proteasomal degradation of TP53wt. Additionally, the use of drugs, such as eprenetapopt, which have been shown to restore TP53wt levels, could be a promising approach [65]
The identification of TP53 as a key molecular marker and driver of BL has important implications for diagnosis, prognosis, and treatment. TP53 testing should thus be incorporated into routine clinical practice to stratify patients according to their risk and response to therapy. Moreover, novel therapeutic strategies that target TP53 or its regulators may offer new opportunities to improve the outcomes of BL patients, especially those with relapsed or refractory disease.
In addition to TP53 abnormalities, other factors that can affect the TP53 pathway in BL include the expression of MDM2 and MDM4, two negative regulators of TP53 that can bind to and inhibit its activity. MDM2 overexpression has been reported in some cases of BL, especially those with immunodeficiency or double-hit status [64][66]. MDM4 overexpression has also been identified as the only TP53 pathway abnormality in a subset of BL cases without TP53 or MDM2 abnormalities, suggesting a potential alternative mechanism of TP53 inhibition [64].
2.2.2. TCF3 and ID3 Mutations in BL
Schmitz et al. [60] found mutations in transcription factor 3 (TCF3) in BL. TCF3 is expressed at high levels during B-cell development and is inhibited by the helix-loop-helix protein ID3. Another gene shown to be mutated in BL is cell cycle regulating Cyclin D3 (CCND3), which is a direct target of TCF3, refs. [60][67][68] confirmed these mutations in an independent study, analyzing their frequency and clinical relevance. Their findings revealed that CCND3 and ID3 mutations are correlated with an advanced stage of the disease, representing secondary hits after MYC deregulation, and are essential for lymphomagenesis. Mutations in ID3 have been identified in a subset of BL cases and are thought to contribute to the development of the disease by inhibiting B-cell differentiation and promoting cell proliferation [67]
In a groundbreaking study, ref. [22] sequenced the genome of a BL tumor and the germline DNA from the same individual, as well as the exomes of over 50 BL tumors, comparing them to diffuse large B-cell lymphoma (DLBCL) tumor samples. They found differentially mutated genes in BL samples, such as ID3, PIK3R1, RET, GNA13, ARID1A, and SMARCA4. Importantly, they identified ID3 mutations occurring specifically in Burkitt lymphoma samples (34% of tumors) and not in DLBCLs. The study was the first to show that ID3 mutations are responsible for promoting cell cycle progression and proliferation in Burkitt lymphoma and identifying ID3 as a new tumor suppressor gene.
2.2.3. MYC Translocation and 11q Alterations
Notably, MYC translocation serves as both a diagnostic hallmark and a critical prognostic factor for BL [69]. Patients afflicted with MYC-positive lymphomas face poorer prognoses and necessitate more aggressive treatment regimens compared to their MYC-negative counterparts [26][70] MYC-negative lymphomas, such as those harboring 11q alterations, are associated with a subtype of high-grade B-cell lymphoma, previously referred to as Burkitt-like lymphoma [71]. The lymphomas with 11q aberrations do not have the characteristic MYC translocations seen in Burkitt lymphoma. Instead, they exhibit chromosomal abnormalities involving the loss of a region on the long arm of chromosome 11 (11q). This region contains several tumor suppressor genes, such as ATM and BIRC3, which play roles in DNA repair and apoptosis. Loss of these tumor suppressor genes can contribute to the development of lymphoma. Although both MYC-positive and -negative subtypes share some genomic features, the presence or absence of MYC translocations and 11q alterations help in distinguishing these two distinct molecular subtypes of high-grade B-cell lymphomas. The grim prognosis of MYC-positive lymphomas (such as BL) is predominantly attributed to the intensified cellular proliferation and genetic instability driven by MYC’s activation of target genes governing cell cycle regulation, DNA replication, and DNA repair [72]. This genomic destabilization ultimately fosters the accumulation of additional mutations, accelerating disease progression and heightening resistance to treatment.
2.3. Evolutionary Growth Advantage as an Implication of MYC Translocation in Cancer
Multiple studies have investigated the role of MYC translocation in providing an evolutionary growth advantage to Burkitt lymphoma (BL) cells. Mlynarczyk et al. [73] proposed that BL cells exploit the germinal center reaction to gain a competitive advantage. This complex process generates high-affinity B-cell receptors through somatic hypermutation and selection. MYC translocation drives the expansion of BL cells within the germinal center, promoting their survival and proliferation, facilitated by the interaction with the microenvironment, including T-cells and stromal cells [73]
Epigenetic regulation of MYC in BL expression and progression has also been the focus of many studies [74][75][76][77]. Fernández-Serrano et al. [78] reviewed the role of epigenetic modifications, such as histone modifications H3K4me3 and H3K27ac, in regulating MYC expression in BL cells. Li et al. [79] demonstrated that MYC mutations in lymphomas are associated with changes in chromatin structure, increased levels of H3K27ac histone marks, and altered gene expression patterns, promoting lymphoma cell proliferation and survival. Cowling et al. [80] reported enhanced histone acetylation of MYC-target genes in lymphomas. Ref. [81] investigated the role of epigenetic mechanisms in MYC-driven lymphomagenesis, revealing alterations in DNA methylation patterns and their impact on specific genes involved in immunological regulation, cell adhesion, and cancer-related pathways; overall indicating a MYC-dominated growth advantage in these cells.
Recent studies have also explored the critical role of microRNAs (miRNAs) and mutations in MYC and its translocated partner genes. Ref. [82] identified several microRNAs that regulate MYC expression, such as miR-150, miR-155, and miR-143, whose deregulation in BL leads to MYC overexpression. Several miRNAs, including miR-34a, miR-145, and the let-7 family, have also been shown to directly target MYC mRNA and negatively regulate its expression [83]. On the other hand, it has also been suggested that c-MYC modulates miRNA expression, revealing a complex interplay between c-MYC and miRNAs [84][85]. One study found 211 differentially expressed genes and 49 differentially expressed miRNAs in BL [86]. A significant finding was the downregulation of ATM and NLK genes, important regulators in response to DNA damage in BL tumor cells. These tumor suppressors were targeted by multiple upregulated miRNAs which could account for their aberrant expression in BL. The combined loss of p53 induction and function due to miRNA-mediated regulation of ATM and NLK, along with the upregulation of TFAP4, may be central for human miRNAs in BL oncogenesis. This allows for the survival of BL tumor cells with the IgH-MYC chromosomal translocation and promotes MYC-induced cell cycle progression, initiating BL lymphomagenesis [86].
Klanova and Klener [87] found that mutations in MYC-translocated partner gene, BCL2, promote the growth and survival of lymphoma cells by increasing the expression of MYC and other oncogenes. MYC mutations are also found to impact the tumor microenvironment, a determining factor for chemotherapeutic potency [88][89][90] These studies showed that MYC mutations in lymphomas lead to increased recruitment of immune-suppressive cells, promoting tumor growth, angiogenesis, and metastasis.
References
- Epstein, M.A.; Achong, B.G.; Barr, Y.M. Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1964, 1, 702–703.
- Zech, L.; Haglund, U.; Nilsson, K.; Klein, G. Characteristic chromosomal abnormalities in biopsies and lymphoid-cell lines from patients with Burkitt and non-Burkitt lymphomas. Int. J. Cancer 1976, 17, 47–56.
- Schulz, T.F.; Boshoff, C.H.; Weiss, R.A. HIV infection and neoplasia. Lancet 1996, 348, 587–591.
- Burkitt, D.P. Etiology of Burkitt’s lymphoma—An alternative hypothesis to a vectored virus. J. Natl. Cancer Inst. 1969, 42, 19–28.
- Parkin, D.M.; Ferlay, J.; Hamdi-Chérif, M.; Sitas, F.; Thomas, J.O.; Wabinga, H.; Whelan, S.L. Cancer in Africa: Epidemiology and Prevention; IARC Press: Lyon, France, 2003.
- Orem, J.; Mbidde, E.K.; Lambert, B.; de Sanjose, S.; Weiderpass, E. Burkitt’s lymphoma in Africa, a review of the epidemiology and etiology. Afr. Health Sci. 2007, 7, 166–175.
- Chang, D.W.; Claassen, G.F.; Hann, S.R.; Cole, M.D. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol. Cell. Biol. 2000, 20, 4309–4319.
- Chen, H.; Liu, H.; Qing, G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 2018, 3, 5.
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC oncogene—The grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2022, 19, 23–36.
- Kress, T.R.; Sabò, A.; Amati, B. MYC: Connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer 2015, 15, 593–607.
- Kalkat, M.; De Melo, J.; Hickman, K.A.; Lourenco, C.; Redel, C.; Resetca, D.; Tamachi, A.; Tu, W.B.; Penn, L.Z. MYC Deregulation in Primary Human Cancers. Genes 2017, 8, 151.
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905.
- Boxer, L.M.; Dang, C.V. Translocations involving c-myc and c-myc function. Oncogene 2001, 20, 5595–5610.
- Pelengaris, S.; Khan, M. The many faces of c-MYC. Arch. Biochem. Biophys. 2003, 416, 129–136.
- Bettess, M.D.; Dubois, N.; Murphy, M.J.; Dubey, C.; Roger, C.; Robine, S.; Trumpp, A. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol. Cell. Biol. 2005, 25, 7868–7878.
- De la Cova, C.; Abril, M.; Bellosta, P.; Gallant, P.; Johnston, L.A. Drosophila myc regulates organ size by inducing cell competition. Cell 2004, 117, 107–116.
- Shi, Y.; Glynn, J.M.; Guilbert, L.J.; Cotter, T.G.; Bissonnette, R.P.; Green, D.R. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science 1992, 257, 212–214.
- Kaczmarek, L.; Hyland, J.K.; Watt, R.; Rosenberg, M.; Baserga, R. Microinjected c-myc as a competence factor. Science 1985, 228, 1313–1315.
- Johnston, J.M.; Carroll, W.L. c-myc hypermutation in Burkitt’s lymphoma. Leuk. Lymphoma 1992, 8, 431–439.
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35.
- Vecchio, E.; Fiume, G.; Correnti, S.; Romano, S.; Iaccino, E.; Mimmi, S.; Maisano, D.; Nisticò, N.; Quinto, I. Insights about MYC and Apoptosis in B-Lymphomagenesis: An Update from Murine Models. Int. J. Mol. Sci. 2020, 21, 4265.
- Love, C.; Sun, Z.; Jima, D.; Li, G.; Zhang, J.; Miles, R.; Richards, K.L.; Dunphy, C.H.; Choi, W.W.; Srivastava, G.; et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 2012, 44, 1321–1325.
- Dalla-Favera, R.; Bregni, M.; Erikson, J.; Patterson, D.; Gallo, R.C.; Croce, C.M. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7824–7827.
- Taub, R.; Kirsch, I.; Morton, C.; Lenoir, G.; Swan, D.; Tronick, S.; Aaronson, S.; Leder, P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7837–7841.
- Hamlyn, P.H.; Rabbitts, T.H. Translocation joins c-myc and immunoglobulin gamma 1 genes in a Burkitt lymphoma revealing a third exon in the c-myc oncogene. Nature 1983, 304, 135–139.
- Nguyen, L.; Papenhausen, P.; Shao, H. The Role of c-MYC in B-Cell Lymphomas: Diagnostic and Molecular Aspects. Genes 2017, 8, 116.
- Basso, K.; Frascella, E.; Zanesco, L.; Rosolen, A. Improved long-distance polymerase chain reaction for the detection of t(8;14)(q24;q32) in Burkitt’s lymphomas. Am. J. Pathol. 1999, 155, 1479–1485.
- Blum, K.A.; Lozanski, G.; Byrd, J.C. Adult Burkitt leukemia and lymphoma. Blood 2004, 104, 3009–3020.
- Hecht, J.L.; Aster, J.C. Molecular biology of Burkitt’s lymphoma. J. Clin. Oncol. 2000, 18, 3707–3721.
- Neri, A.; Barriga, F.; Knowles, D.M.; Magrath, I.T.; Dalla-Favera, R. Different regions of the immunoglobulin heavy-chain locus are involved in chromosomal translocations in distinct pathogenetic forms of Burkitt lymphoma. Proc. Natl. Acad. Sci. USA 1988, 85, 2748–2752.
- Devaiah, B.N.; Mu, J.; Akman, B.; Uppal, S.; Weissman, J.D.; Cheng, D.; Baranello, L.; Nie, Z.; Levens, D.; Singer, D.S. MYC protein stability is negatively regulated by BRD4. Proc. Natl. Acad. Sci. USA 2020, 117, 13457–13467.
- Hinds, J.W.; Feris, E.J.; Wilkins, O.M.; Deary, L.T.; Wang, X.; Cole, M.D. S146L in MYC is a context-dependent activating substitution in cancer development. PLoS ONE 2022, 17, e0272771.
- Kuzyk, A.; Mai, S. c-MYC-induced genomic instability. Cold Spring Harb. Perspect. Med. 2014, 4, a014373.
- Kumari, A.; Folk, W.P.; Sakamuro, D. The Dual Roles of MYC in Genomic Instability and Cancer Chemoresistance. Genes 2017, 8, 158.
- Curti, L.; Campaner, S. MYC-Induced Replicative Stress: A Double-Edged Sword for Cancer Development and Treatment. Int. J. Mol. Sci. 2021, 22, 6168.
- Shortt, J.; Martin, B.P.; Newbold, A.; Hannan, K.M.; Devlin, J.R.; Baker, A.J.; Ralli, R.; Cullinane, C.; Schmitt, C.A.; Reimann, M.; et al. Combined inhibition of PI3K-related DNA damage response kinases and mTORC1 induces apoptosis in MYC-driven B-cell lymphomas. Blood 2013, 121, 2964–2974.
- Han, S.S.; Yun, H.; Son, D.J.; Tompkins, V.S.; Peng, L.; Chung, S.T.; Kim, J.S.; Park, E.S.; Janz, S. NF-kappaB/STAT3/PI3K signaling crosstalk in iMyc E mu B lymphoma. Mol. Cancer 2010, 9, 97.
- Dang, C.V. The interplay between MYC and HIF in the Warburg effect. In Ernst Schering Foundation Symposium Proceedings; Springer: Berlin/Heidelberg, Germany, 2007; pp. 35–53.
- Dang, C.V. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a014217.
- Kim, J.; Lee, J.H.; Iyer, V.R. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS ONE 2008, 3, e1798.
- Popay, T.M.; Wang, J.; Adams, C.M.; Howard, G.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A.; Thomas, L.R.; Lorey, S.L.; Machida, Y.J.; et al. MYC regulates ribosome biogenesis and mitochondrial gene expression programs through its interaction with host cell factor-1. eLife 2021, 10, e60191.
- Morrish, F.; Hockenbery, D. MYC and mitochondrial biogenesis. Cold Spring Harb. Perspect. Med. 2014, 4, a014225.
- Casey, S.C.; Baylot, V.; Felsher, D.W. The MYC oncogene is a global regulator of the immune response. Blood 2018, 131, 2007–2015.
- Casey, S.C.; Tong, L.; Li, Y.; Do, R.; Walz, S.; Fitzgerald, K.N.; Gouw, A.M.; Baylot, V.; Gütgemann, I.; Eilers, M.; et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016, 352, 227–231.
- Kim, E.Y.; Kim, A.; Kim, S.K.; Chang, Y.S. MYC expression correlates with PD-L1 expression in non-small cell lung cancer. Lung Cancer 2017, 110, 63–67.
- Saravia, J.; Zeng, H.; Dhungana, Y.; Bastardo Blanco, D.; Nguyen, T.M.; Chapman, N.M.; Wang, Y.; Kanneganti, A.; Liu, S.; Raynor, J.L.; et al. Homeostasis and transitional activation of regulatory T cells require c-Myc. Sci. Adv. 2020, 6, eaaw6443.
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401.
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1.
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990.
- Stasevich, E.M.; Uvarova, A.N.; Murashko, M.M.; Khabusheva, E.R.; Sheetikov, S.A.; Prassolov, V.S.; Kuprash, D.V.; Demin, D.E.; Schwartz, A.M. Enhancer RNA AL928768.3 from the IGH Locus Regulates MYC Expression and Controls the Proliferation and Chemoresistance of Burkitt Lymphoma Cells with IGH/MYC Translocation. Int. J. Mol. Sci. 2022, 23, 4624.
- Brady, G.; MacArthur, G.J.; Farrell, P.J. Epstein-Barr virus and Burkitt lymphoma. J. Clin. Pathol. 2007, 60, 1397–1402.
- Broen, K.; Dickens, J.; Trangucci, R.; Ogwang, M.D.; Tenge, C.N.; Masalu, N.; Reynolds, S.J.; Kawira, E.; Kerchan, P.; Were, P.A.; et al. Burkitt lymphoma risk shows geographic and temporal associations with Plasmodium falciparum infections in Uganda, Tanzania, and Kenya. Proc. Natl. Acad. Sci. USA 2023, 120, e2211055120.
- Blinder, V.S.; Chadburn, A.; Furman, R.R.; Mathew, S.; Leonard, J.P. Improving outcomes for patients with Burkitt lymphoma and HIV. AIDS Patient Care STDs 2008, 22, 175–187.
- Bemark, M.; Neuberger, M.S. The c-MYC allele that is translocated into the IgH locus undergoes constitutive hypermutation in a Burkitt’s lymphoma line. Oncogene 2000, 19, 3404–3410.
- Schmitz, R.; Ceribelli, M.; Pittaluga, S.; Wright, G.; Staudt, L.M. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb. Perspect. Med. 2014, 4, a014282.
- Hummel, M.; Bentink, S.; Berger, H.; Klapper, W.; Wessendorf, S.; Barth, T.F.; Bernd, H.W.; Cogliatti, S.B.; Dierlamm, J.; Feller, A.C.; et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N. Engl. J. Med. 2006, 354, 2419–2430.
- López, C.; Kleinheinz, K.; Aukema, S.M.; Rohde, M.; Bernhart, S.H.; Hübschmann, D.; Wagener, R.; Toprak, U.H.; Raimondi, F.; Kreuz, M.; et al. Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma. Nat. Commun. 2019, 10, 1459.
- Küppers, R.; Dalla-Favera, R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene 2001, 20, 5580–5594.
- Tandon, A.; Birkenhagen, J.; Nagalla, D.; Kölker, S.; Sauer, S.W. ADP-dependent glucokinase as a novel onco-target for haematological malignancies. Sci. Rep. 2020, 10, 13584.
- Schmitz, R.; Young, R.M.; Ceribelli, M.; Jhavar, S.; Xiao, W.; Zhang, M.; Wright, G.; Shaffer, A.L.; Hodson, D.J.; Buras, E.; et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 2012, 490, 116–120.
- Sánchez-Beato, M.; Sánchez-Aguilera, A.; Piris, M.A. Cell cycle deregulation in B-cell lymphomas. Blood 2003, 101, 1220–1235.
- Newman, A.M.; Zaka, M.; Zhou, P.; Blain, A.E.; Erhorn, A.; Barnard, A.; Crossland, R.E.; Wilkinson, S.; Enshaei, A.; De Zordi, J.; et al. Genomic abnormalities of TP53 define distinct risk groups of paediatric B-cell non-Hodgkin lymphoma. Leukemia 2022, 36, 781–789.
- López, C.; Burkhardt, B.; Chan, J.K.C.; Leoncini, L.; Mbulaiteye, S.M.; Ogwang, M.D.; Orem, J.; Rochford, R.; Roschewski, M.; Siebert, R. Burkitt lymphoma. Nat. Rev. Dis. Prim. 2022, 8, 78.
- Leventaki, V.; Rodic, V.; Tripp, S.R.; Bayerl, M.G.; Perkins, S.L.; Barnette, P.; Schiffman, J.D.; Miles, R.R. TP53 pathway analysis in paediatric Burkitt lymphoma reveals increased MDM4 expression as the only TP53 pathway abnormality detected in a subset of cases. Br. J. Haematol. 2012, 158, 763–771.
- Burkhardt, B.; Michgehl, U.; Rohde, J.; Erdmann, T.; Berning, P.; Reutter, K.; Rohde, M.; Borkhardt, A.; Burmeister, T.; Dave, S.; et al. Clinical relevance of molecular characteristics in Burkitt lymphoma differs according to age. Nat. Commun. 2022, 13, 3881.
- Yu, D.; Carroll, M.; Thomas-Tikhonenko, A. p53 status dictates responses of B lymphomas to monotherapy with proteasome inhibitors. Blood 2007, 109, 4936–4943.
- Richter, J.; Schlesner, M.; Hoffmann, S.; Kreuz, M.; Leich, E.; Burkhardt, B.; Rosolowski, M.; Ammerpohl, O.; Wagener, R.; Bernhart, S.H.; et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat. Genet. 2012, 44, 1316–1320.
- Rohde, M.; Bonn, B.R.; Zimmermann, M.; Lange, J.; Möricke, A.; Klapper, W.; Oschlies, I.; Szczepanowski, M.; Nagel, I.; Schrappe, M.; et al. Relevance of ID3-TCF3-CCND3 pathway mutations in pediatric aggressive B-cell lymphoma treated according to the non-Hodgkin Lymphoma Berlin-Frankfurt-Münster protocols. Haematologica 2017, 102, 1091–1098.
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390.
- Lin, P.; Dickason, T.J.; Fayad, L.E.; Lennon, P.A.; Hu, P.; Garcia, M.; Routbort, M.J.; Miranda, R.; Wang, X.; Qiao, W.; et al. Prognostic value of MYC rearrangement in cases of B-cell lymphoma, unclassifiable, with features intermediate between diffuse large B-cell lymphoma and Burkitt lymphoma. Cancer 2012, 118, 1566–1573.
- Salaverria, I.; Martin-Guerrero, I.; Wagener, R.; Kreuz, M.; Kohler, C.W.; Richter, J.; Pienkowska-Grela, B.; Adam, P.; Burkhardt, B.; Claviez, A.; et al. A recurrent 11q aberration pattern characterizes a subset of MYC-negative high-grade B-cell lymphomas resembling Burkitt lymphoma. Blood 2014, 123, 1187–1198.
- García-Gutiérrez, L.; Delgado, M.D.; León, J. MYC Oncogene Contributions to Release of Cell Cycle Brakes. Genes 2019, 10, 244.
- Mlynarczyk, C.; Fontán, L.; Melnick, A. Germinal center-derived lymphomas: The darkest side of humoral immunity. Immunol. Rev. 2019, 288, 214–239.
- Lindström, M.S.; Wiman, K.G. Role of genetic and epigenetic changes in Burkitt lymphoma. Semin. Cancer Biol. 2002, 12, 381–387.
- McMahon, S.B.; Wood, M.A.; Cole, M.D. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 2000, 20, 556–562.
- Park, J.; Kunjibettu, S.; McMahon, S.B.; Cole, M.D. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 2001, 15, 1619–1624.
- Ribeiro, M.L.; Reyes-Garau, D.; Armengol, M.; Fernández-Serrano, M.; Roué, G. Recent Advances in the Targeting of Epigenetic Regulators in B-Cell Non-Hodgkin Lymphoma. Front. Genet. 2019, 10, 986.
- Fernández-Serrano, M.; Winkler, R.; Santos, J.C.; Le Pannérer, M.M.; Buschbeck, M.; Roué, G. Histone Modifications and Their Targeting in Lymphoid Malignancies. Int. J. Mol. Sci. 2021, 23, 253.
- Li, Z.; Van Calcar, S.; Qu, C.; Cavenee, W.K.; Zhang, M.Q.; Ren, B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc. Natl. Acad. Sci. USA 2003, 100, 8164–8169.
- Cowling, V.H.; Turner, S.A.; Cole, M.D. Burkitt’s lymphoma-associated c-Myc mutations converge on a dramatically altered target gene response and implicate Nol5a/Nop56 in oncogenesis. Oncogene 2014, 33, 3519–3527.
- Poole, C.J.; Zheng, W.; Lodh, A.; Yevtodiyenko, A.; Liefwalker, D.; Li, H.; Felsher, D.W.; van Riggelen, J. DNMT3B overexpression contributes to aberrant DNA methylation and MYC-driven tumor maintenance in T-ALL and Burkitt’s lymphoma. Oncotarget 2017, 8, 76898–76920.
- Videtta, A.D.; Malagnino, V.; De Falco, G. Current understanding of the role and regulation of miRNAs in Burkitt lymphoma. Blood Lymphat. Cancer Targets Ther. 2018, 8, 33–45.
- Shams, R.; Asadzadeh Aghdaei, H.; Behmanesh, A.; Sadeghi, A.; Zali, M.; Salari, S.; Padrón, J.M. MicroRNAs Targeting MYC Expression: Trace of Hope for Pancreatic Cancer Therapy. A Systematic Review. Cancer Manag. Res. 2020, 12, 2393–2404.
- Onnis, A.; De Falco, G.; Antonicelli, G.; Onorati, M.; Bellan, C.; Sherman, O.; Sayed, S.; Leoncini, L. Alteration of microRNAs regulated by c-Myc in Burkitt lymphoma. PLoS ONE 2010, 5, e12960.
- Bui, T.V.; Mendell, J.T. Myc: Maestro of microRNAs. Genes Cancer 2010, 1, 568–575.
- Oduor, C.I.; Kaymaz, Y.; Chelimo, K.; Otieno, J.A.; Ong’echa, J.M.; Moormann, A.M.; Bailey, J.A. Integrative microRNA and mRNA deep-sequencing expression profiling in endemic Burkitt lymphoma. BMC Cancer 2017, 17, 761.
- Klanova, M.; Klener, P. BCL-2 Proteins in Pathogenesis and Therapy of B-Cell Non-Hodgkin Lymphomas. Cancers 2020, 12, 938.
- Meškytė, E.M.; Keskas, S.; Ciribilli, Y. MYC as a Multifaceted Regulator of Tumor Microenvironment Leading to Metastasis. Int. J. Mol. Sci. 2020, 21, 7710.
- Dews, M.; Homayouni, A.; Yu, D.; Murphy, D.; Sevignani, C.; Wentzel, E.; Furth, E.E.; Lee, W.M.; Enders, G.H.; Mendell, J.T.; et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006, 38, 1060–1065.
- Whitfield, J.R.; Soucek, L. Tumor microenvironment: Becoming sick of Myc. Cell. Mol. Life Sci. 2012, 69, 931–934.
More
Information
Subjects:
Immunology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
567
Revisions:
2 times
(View History)
Update Date:
20 Jul 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





