
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Xueyan Wang | -- | 2689 | 2023-07-04 09:13:02 | | | |
| 2 | Dean Liu | -24 word(s) | 2665 | 2023-07-05 02:09:15 | | |
Video Upload Options
The emergence of precision biology has been driven by the development of advanced technologies and techniques in high-resolution biological research systems. Enhancer-mediated transcriptional regulation, a complex network of gene expression and regulation in eukaryotes, has attracted significant attention as a promising avenue for investigating the underlying mechanisms of biological processes and diseases. To address biological problems with precision, large amounts of data, functional information, and research on the mechanisms of action of biological molecules is required to address biological problems with precision. Enhancers, including typical enhancers and super enhancers, play a crucial role in gene expression and regulation within this network. The identification and targeting of disease-associated enhancers hold the potential to advance precision medicine.
1. Introduction
2. The Development of Precision Biology
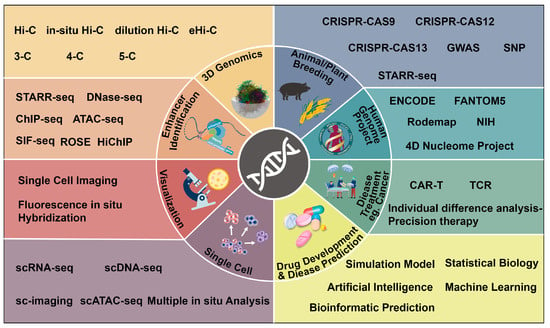
3. Transcriptional Regulation and Precision Biology
4. The Role of Different Enhancers in Transcriptional Regulation
4.1. Typical Enhancers
4.2. Annotation of Typical Enhancers
4.3. Super Enhancer
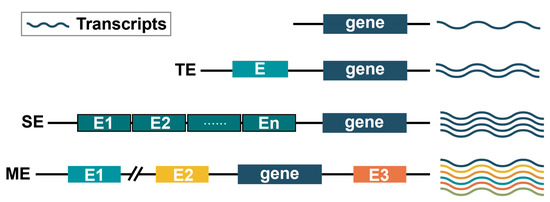
4.4. Multi-Enhancer Modulation
References
- Weil, A.R. Precision Medicine. Health Aff. 2018, 37, 687.
- Nakagawa, H.; Fujita, M. Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci. 2018, 109, 513–522.
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.-P.; Simon, P.; Löwer, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrörs, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226.
- Sookoian, S.; Pirola, C.J. Precision medicine in nonalcoholic fatty liver disease: New therapeutic insights from genetics and systems biology. Clin. Mol. Hepatol. 2020, 26, 461–475.
- Fountzilas, E.; Tsimberidou, A.M.; Vo, H.H.; Kurzrock, R. Clinical trial design in the era of precision medicine. Genome Med. 2022, 14, 101.
- Alaimo, S.; Giugno, R.; Acunzo, M.; Veneziano, D.; Ferro, A.; Pulvirenti, A. Post-transcriptional knowledge in pathway analysis increases the accuracy of phenotypes classification. Oncotarget 2016, 7, 54572–54582.
- Kampmann, M. CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine. ACS Chem. Biol. 2018, 13, 406–416.
- Galis, Z.S. Editorial: Where Is Waldo: Contextualizing the Endothelial Cell in the Era of Precision Biology. Front. Cardiovasc. Med. 2020, 7, 127.
- Stadhouders, R.; Filion, G.J.; Graf, T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature 2019, 569, 345–354.
- Bentovim, L.; Harden, T.; DePace, A.H. Transcriptional precision and accuracy in development: From measurements to models and mechanisms. Development 2017, 144, 3855–3866.
- Zhang, Y.; Wong, C.-H.; Birnbaum, R.Y.; Li, G.; Favaro, R.; Ngan, C.Y.; Lim, J.; Tai, E.; Poh, H.M.; Wong, E.; et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 2013, 504, 306–310.
- Nagai, S.; Davis, R.E.; Mattei, P.J.; Eagen, K.P.; Kornberg, R.D. Chromatin potentiates transcription. Proc. Natl. Acad. Sci. USA 2017, 114, 1536–1541.
- The ENCODE Project Consortium. The ENCODE (ENCyclopedia of DNA Elements) Project. Science 2004, 306, 636–640.
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108.
- Kvon, E.Z.; Kazmar, T.; Stampfel, G.; Yáñez-Cuna, J.O.; Pagani, M.; Schernhuber, K.; Dickson, B.J.; Stark, A. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 2014, 512, 91–95.
- Lee, T.I.; Young, R.A. Transcriptional Regulation and Its Misregulation in Disease. Cell 2013, 152, 1237–1251.
- Bulger, M.; Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 2011, 144, 327–339.
- Li, W.; Notani, D.; Rosenfeld, M.G. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 2016, 17, 207–223.
- Heintzman, N.D.; Hon, G.C.; Hawkins, R.D.; Kheradpour, P.; Stark, A.; Harp, L.F.; Ye, Z.; Lee, L.K.; Stuart, R.K.; Ching, C.W.; et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009, 459, 108–112.
- Pennacchio, L.A.; Bickmore, W.; Dean, A.; Nobrega, M.A.; Bejerano, G. Enhancers: Five essential questions. Nat. Rev. Genet. 2013, 14, 288–295.
- Laiker, I.; Frankel, N. Pleiotropic Enhancers are Ubiquitous Regulatory Elements in the Human Genome. Genome Biol. Evol. 2022, 14, evac071.
- Yang, H.-T.; Shah, R.H.; Tegay, D.; Onel, K. Precision oncology: Lessons learned and challenges for the future. Cancer Manag. Res. 2019, 11, 7525–7536.
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319.
- Hnisz, D.; Schuijers, J.; Lin, C.Y. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell 2015, 58, 362–370.
- Chen, H.; Liang, H. A High-Resolution Map of Human Enhancer RNA Loci Characterizes Super-enhancer Activities in Cancer. Cancer Cell 2020, 38, 701–715.
- Collins, F.S.; Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 2015, 372, 793–795.
- Auffray, C.; Griffin, J.L.; Khoury, M.J.; Lupski, J.R.; Schwab, M. Ten years of Genome Medicine. Genome Med. 2019, 11, 7.
- Goetz, L.H.; Schork, N.J. Personalized medicine: Motivation, challenges, and progress. Fertil. Steril. 2018, 109, 952–963.
- Elemento, O. The future of precision medicine: Towards a more predictive personalized medicine. Emerg. Top. Life Sci. 2020, 4, 175–177.
- Ozdemir, E.S.; Ranganathan, S.V.; Nussinov, R. How has artificial intelligence impacted COVID-19 drug repurposing and what lessons have we learned? Expert Opin. Drug Discov. 2022, 17, 1061–1065.
- Filipp, F.V. Opportunities for Artificial Intelligence in Advancing Precision Medicine. Curr. Genet. Med. Rep. 2019, 7, 208–213.
- Sun, W.; Lee, J.; Zhang, S.; Benyshek, C.; Dokmeci, M.R.; Khademhosseini, A. Engineering Precision Medicine. Adv. Sci. 2019, 6, 1801039.
- Splinter, E.; de Laat, W. The complex transcription regulatory landscape of our genome: Control in three dimensions. EMBO J. 2011, 30, 4345–4355.
- Dostie, J.; Richmond, T.A.; Arnaout, R.A.; Selzer, R.R.; Lee, W.L.; Honan, T.A.; Rubio, E.D.; Krumm, A.; Lamb, J.; Nusbaum, C.; et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006, 16, 1299–1309.
- Rao, S.S.P.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680.
- Nagano, T.; Lubling, Y.; Stevens, T.J.; Schoenfelder, S.; Yaffe, E.; Dean, W.; Laue, E.D.; Tanay, A.; Fraser, P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013, 502, 59–64.
- Kong, S.; Li, Q.; Zhang, G.; Li, Q.; Huang, Q.; Huang, L.; Zhang, H.; Huang, Y.; Peng, Y.; Qin, B.; et al. Exonuclease combinations reduce noises in 3D genomics technologies. Nucleic Acids Res. 2020, 48, e44.
- Li, G.; Fullwood, M.J.; Xu, H.; Mulawadi, F.H.; Velkov, S.; Vega, V.; Ariyaratne, P.N.; Bin Mohamed, Y.; Ooi, H.-S.; Tennakoon, C.; et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010, 11, R22.
- Solomon, M.J.; Larsen, P.L.; Varshavsky, A. Mapping protein-DNA interactions in vivo with formaldehyde: Evidence that histone H4 is retained on a highly transcribed gene. Cell 1988, 53, 937–947.
- Mundade, R.; Ozer, H.G.; Wei, H.; Prabhu, L.; Lu, T. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell Cycle 2014, 13, 2847–2852.
- Yang, Y.; Li, C.; Chen, Z.; Zhang, Y.; Tian, Q.; Sun, M.; Zhang, S.; Yu, M.; Wang, G. An intellectual disability-related MED23 mutation dysregulates gene expression by altering chromatin conformation and enhancer activities. Nucleic Acids Res. 2023, 51, 2137–2150.
- Lin, D.; Xu, W.; Hong, P.; Wu, C.; Zhang, Z.; Zhang, S.; Xing, L.; Yang, B.; Zhou, W.; Xiao, Q.; et al. Decoding the spatial chromatin organization and dynamic epigenetic landscapes of macrophage cells during differentiation and immune activation. Nat. Commun. 2022, 13, 5857.
- Dickel, D.E.; Zhu, Y.; Nord, A.S.; Wylie, J.N.; Akiyama, J.A.; Afzal, V.; Plajzer-Frick, I.; Kirkpatrick, A.; Göttgens, B.; Bruneau, B.G.; et al. Function-based identification of mammalian enhancers using site-specific integration. Nat. Methods 2014, 11, 566–571.
- Liu, S.; Liu, Y.; Zhang, Q.; Wu, J.; Liang, J.; Yu, S.; Wei, G.-H.; White, K.P.; Wang, X. Systematic identification of regulatory variants associated with cancer risk. Genome Biol. 2017, 18, 194.
- Benoit, M. Shooting for the STARRs: A Modified STARR-seq Assay for Rapid Identification and Evaluation of Plant Regulatory Sequences in Tobacco Leaves. Plant Cell 2020, 32, 2057–2058.
- Sbalzarini, I.F.; Koumoutsakos, P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 2005, 151, 182–195.
- Lee, L.P. Satellite nanoscope and cellular BioASICs for quantitative biomedicine. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2009, 2009, 4582–4585.
- Dasgupta, A.; Deschamps, J.; Matti, U.; Hübner, U.; Becker, J.; Strauss, S.; Jungmann, R.; Heintzmann, R.; Ries, J. Direct supercritical angle localization microscopy for nanometer 3D superresolution. Nat. Commun. 2021, 12, 1180.
- Tanay, A.; Regev, A. Scaling single-cell genomics from phenomenology to mechanism. Nature 2017, 541, 331–338.
- Navin, N.E. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015, 25, 1499–1507.
- Chessel, A.; Carazo Salas, R.E. From observing to predicting single-cell structure and function with high-throughput/high-content microscopy. Essays Biochem. 2019, 63, 197–208.
- Lu, T.; Ang, C.E.; Zhuang, X. Spatially resolved epigenomic profiling of single cells in complex tissues. Cell 2022, 185, 4448–4464.
- Kelsey, G.; Stegle, O.; Reik, W. Single-cell epigenomics: Recording the past and predicting the future. Science 2017, 358, 69–75.
- Shema, E.; Bernstein, B.E.; Buenrostro, J.D. Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet. 2019, 51, 19–25.
- Li, J.; Chin, C.R.; Ying, H.Y.; Meydan, C.; Teater, M.R.; Xia, M.; Farinha, P.; Takata, K.; Chu, C.S.; Rivas, M.A.; et al. Cooperative super-enhancer inactivation caused by heterozygous loss of CREBBP and KMT2D skews B cell fate decisions and yields T cell-depleted lymphomas. bioRxiv 2023.
- Cusanovich, D.A.; Caliskan, M.; Billstrand, C.; Michelini, K.; Chavarria, C.; De Leon, S.; Mitrano, A.; Lewellyn, N.; Elias, J.A.; Chupp, G.L.; et al. Integrated analyses of gene expression and genetic association studies in a founder population. Hum. Mol. Genet. 2016, 25, 2104–2112.
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020, 21, 71–87.
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945.
- Dekker, J.; Belmont, A.S.; Guttman, M.; Leshyk, V.O.; Lis, J.T.; Lomvardas, S.; Mirny, L.A.; O’shea, C.C.; Park, P.J.; Ren, B.; et al. The 4D nucleome project. Nature 2017, 549, 219–226.
- Zhou, X.; Lu, Y.; Zhao, F.; Dong, J.; Ma, W.; Zhong, S.; Wang, M.; Wang, B.; Zhao, Y.; Shi, Y.; et al. Deciphering the spatial-temporal transcriptional landscape of human hypothalamus development. Cell Stem Cell 2022, 29, 328–343.
- Banerji, J.; Rusconi, S.; Schaffner, W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 1981, 27, 299–308.
- Moreau, P.; Hen, R.; Wasylyk, B.; Everett, R.; Gaub, M.; Chambon, P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981, 9, 6047–6068.
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837.
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; Van Calcar, S.; Qu, C.; Ching, K.A.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007, 39, 311–318.
- Calo, E.; Wysocka, J. Modification of enhancer chromatin: What, how, and why? Mol. Cell 2013, 49, 825–837.
- Spitz, F.; Furlong, E.E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626.
- Shlyueva, D.; Stampfel, G.; Stark, A. Transcriptional enhancers: From properties to genome-wide predictions. Nat. Rev. Genet. 2014, 15, 272–286.
- Rivera, C.M.; Ren, B. Mapping human epigenomes. Cell 2013, 155, 39–55.
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74.
- Bernstein, B.E.; Stamatoyannopoulos, J.A.; Costello, J.F.; Ren, B.; Milosavljevic, A.; Meissner, A.; Kellis, M.; Marra, M.A.; Beaudet, A.L.; Ecker, J.R.; et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010, 28, 1045–1048.
- Andersson, R.; Gebhard, C.; Miguel-Escalada, I.; Hoof, I.; Bornholdt, J.; Boyd, M.; Chen, Y.; Zhao, X.; Schmidl, C.; Suzuki, T.; et al. An atlas of active enhancers across human cell types and tissues. Nature 2014, 507, 455–461.
- Forrest, A.R.R.; Kawaji, H.; Rehli, M.; Kenneth Baillie, J.; de Hoon, M.J.L.; Haberle, V.; Lassmann, T.; Kulakovskiy, I.V.; Lizio, M.; Itoh, M.; et al. A promoter-level mammalian expression atlas. Nature 2014, 507, 462–470.
- Fernández, J.M.; de la Torre, V.; Richardson, D.; Royo, R.; Puiggròs, M.; Moncunill, V.; Fragkogianni, S.; Clarke, L.; Flicek, P.; Rico, D.; et al. The BLUEPRINT Data Analysis Portal. Cell Syst. 2016, 3, 491–495.e495.
- Visel, A.; Minovitsky, S.; Dubchak, I.; Pennacchio, L.A. VISTA Enhancer Browser–A database of tissue-specific human enhancers. Nucleic Acids Res. 2007, 35, D88–D92.
- Gao, T.; He, B.; Liu, S.; Zhu, H.; Tan, K.; Qian, J. EnhancerAtlas: A resource for enhancer annotation and analysis in 105 human cell/tissue types. Bioinformatics 2016, 32, 3543–3551.
- Wang, Z.; Zhang, Q.; Zhang, W.; Lin, J.-R.; Cai, Y.; Mitra, J.; Zhang, Z.D. HEDD: Human Enhancer Disease Database. Nucleic Acids Res. 2018, 46, D113–D120.
- Visel, A.; Blow, M.J.; Li, Z.; Zhang, T.; Akiyama, J.A.; Holt, A.; Plajzer-Frick, I.; Shoukry, M.; Wright, C.; Chen, F.; et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 2009, 457, 854–858.
- Bonn, S.; Zinzen, R.; Girardot, C.; Gustafson, E.H.; Gonzalez, A.P.; Delhomme, N.; Ghavi-Helm, Y.; Wilczynski, B.; Riddell, A.; Furlong, E. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012, 44, 148–156.
- Pekowska, A.; Benoukraf, T.; Zacarias-Cabeza, J.; Belhocine, M.; Koch, F.; Holota, H.; Imbert, J.; Andrau, J.-C.; Ferrier, P.; Spicuglia, S. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011, 30, 4198–4210.
- Peng, Y.; Zhang, Y. Enhancer and super-enhancer: Positive regulators in gene transcription. Anim. Model Exp. Med. 2018, 1, 169–179.
- Zhang, J.; Yue, W.; Zhou, Y.; Liao, M.; Chen, X.; Hua, J. Super enhancers-Functional cores under the 3D genome. Cell Prolif. 2021, 54, e12970.
- Thomas, H.F.; Kotova, E.; Jayaram, S.; Pilz, A.; Romeike, M.; Lackner, A.; Penz, T.; Bock, C.; Leeb, M.; Halbritter, F.; et al. Temporal dissection of an enhancer cluster reveals distinct temporal and functional contributions of individual elements. Mol. Cell 2021, 81, 969–982.
- Jiang, Y.; Jiang, Y.-Y.; Lin, D.-C. Super-enhancer-mediated core regulatory circuitry in human cancer. Comput. Struct. Biotechnol. J. 2021, 19, 2790–2795.
- Tang, F.; Yang, Z.; Tan, Y.; Li, Y. Super-enhancer function and its application in cancer targeted therapy. NPJ Precis Oncol. 2020, 4, 2.
- Erokhin, M.; Vassetzky, Y.; Georgiev, P.; Chetverina, D. Eukaryotic enhancers: Common features, regulation, and participation in diseases. Cell. Mol. Life Sci. 2015, 72, 2361–2375.
- Chen, F.X.; Xie, P.; Collings, C.K.; Cao, K.; Aoi, Y.; Marshall, S.A.; Rendleman, E.J.; Ugarenko, M.; Ozark, P.A.; Zhang, A.; et al. PAF1 regulation of promoter-proximal pause release via enhancer activation. Science 2017, 357, 1294–1298.
- Chen, F.X.; Woodfin, A.R.; Gardini, A.; Rickels, R.A.; Marshall, S.A.; Smith, E.R.; Shiekhattar, R.; Shilatifard, A. PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell 2015, 162, 1003–1015.




