Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Mara Schvarzstein | -- | 1573 | 2023-06-30 07:30:18 | | | |
| 2 | Wendy Huang | Meta information modification | 1573 | 2023-06-30 13:04:17 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Schvarzstein, M.; Alam, F.; Toure, M.; Yanowitz, J.L. Whole Genome Duplication in Development, Evolution, and Disease. Encyclopedia. Available online: https://encyclopedia.pub/entry/46253 (accessed on 07 February 2026).
Schvarzstein M, Alam F, Toure M, Yanowitz JL. Whole Genome Duplication in Development, Evolution, and Disease. Encyclopedia. Available at: https://encyclopedia.pub/entry/46253. Accessed February 07, 2026.
Schvarzstein, Mara, Fatema Alam, Muhammad Toure, Judith L. Yanowitz. "Whole Genome Duplication in Development, Evolution, and Disease" Encyclopedia, https://encyclopedia.pub/entry/46253 (accessed February 07, 2026).
Schvarzstein, M., Alam, F., Toure, M., & Yanowitz, J.L. (2023, June 30). Whole Genome Duplication in Development, Evolution, and Disease. In Encyclopedia. https://encyclopedia.pub/entry/46253
Schvarzstein, Mara, et al. "Whole Genome Duplication in Development, Evolution, and Disease." Encyclopedia. Web. 30 June, 2023.
Copy Citation
Whole genome duplication (WGD) or polyploidization can occur at the cellular, tissue, and organismal levels. At the cellular level, tetraploidization has been proposed as a driver of aneuploidy and genome instability and correlates strongly with cancer progression, metastasis, and the development of drug resistance. WGD is also a key developmental strategy for regulating cell size, metabolism, and cellular function. In specific tissues, WGD is involved in normal development (e.g., organogenesis), tissue homeostasis, wound healing, and regeneration. At the organismal level, WGD propels evolutionary processes such as adaptation, speciation, and crop domestication.
whole genome duplication
polyploidization
polyploidy
aneuploidy
adaptation
1. Introduction
Whole genome duplication (WGD) or polyploidization is vital for normal development [1], tissue homeostasis [2][3], and regeneration [4] (Figure 1). It is also a driver of evolutionary processes [5] including adaptation [6], speciation [7], and crop domestication [8]. In addition, WGD can drive pathological conditions, especially oncogenesis [1][9][10]. In nature, WGD exists at the cellular, tissue, organ, and organismal levels. Polyploid cells are normally found in most multicellular diploid organisms [11][12]. In humans, certain cells from the placenta [13], mammary glands [14], liver [15], heart [16], skin [17], and bone marrow must become polyploid to perform specialized functions [18], for tissue homeostasis [3], or as part of the wound healing process [19]. For instance, during development, megakaryocytes acquire 16 copies of the genome and thus become giant cells that are hypermetabolic. This polyploidization is a basic requirement to produce platelets in the blood, as platelets are produced by fragmenting the cytoplasm of megakaryocytes [20][21][22]. In addition, WGD is involved in oncogenesis, metastasis, and the development of resistance to chemotherapeutic drugs, and polyploid cells may accumulate in patients with neurodegenerative disorders, cardiovascular disease, and diabetes [23][24][25][26].
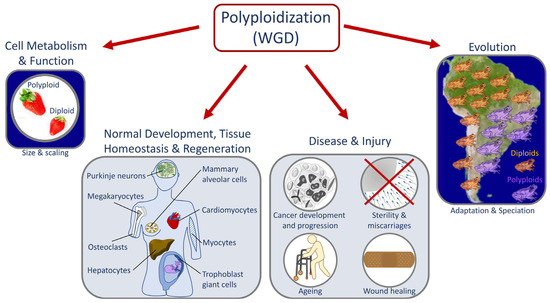
Figure 1. Roles of whole genome duplication (WGD). Diagram depicting key examples of processes affected by polyploidization/WGD. Polyploidy can occur at the cellular, tissue, or organismal levels. It is a key step in important biological processes affecting cellular metabolism and function, normal development, tissue homeostasis, regeneration, wound healing, organismal adaptation to the environment, and speciation. In addition, polyploidization can cause and/or prevent disease and repair injuries.
Polyploid cells can arise by cell fusion resulting in larger multinucleate cells, by endoduplication (alternating cycles between G and S phases without undergoing mitosis or cytokinesis) resulting in mononucleated cells, or by endomitosis (the cell enters mitosis but does not complete cell division) to generate either mononucleated giant cells or multinucleated cells [27]. In some cyanobacteria, polyploidy is established and maintained by misregulation of the replisome machinery [28][29][30].
2. Causes and Downstream Effects of WGD
The outcomes of WGD are diverse in nature—ranging from changes in gene expression to altered cell size and scaling and to adaptation to stress [31][32] (Figure 2). The effects of polyploidization are surprisingly similar in different biological contexts (e.g., in cells, tissues, and organisms) [4][11][33][34] and regardless of the biological process (e.g., during development, evolution, regeneration, and oncogenesis) [1][3][35][36][37][38][39][40][41] where WGD takes place (Figure 2). A common feature of WGD is that it is induced by stress, and it provides the changes that result in plasticity and thus enhances adaptation to stress in normal and pathological conditions. For instance, environmental stresses induce WGD in tissues (e.g., wounding or viral infection) [42][43][44] or organisms (e.g., large changes in temperature or water availability and salinity) [45][46][47][48][49][50]. Furthermore, stress-induced WGD is a platform for genomic diversity and versatility that provides the means for rapid adaptability (Figure 1 and Figure 2). Whereas in whole organisms, this can elicit a competitive advantage in a changing environment [32][51]; in cancer, it elicits an adaptive advantage that promotes oncogenesis, metastasis, and the development of drug resistance [32][51]. The latter is similar to how the pathogenic yeast Candida abdicans becomes resistant to antifungal drugs [6]. Therefore, it is widely expected that queries about mechanisms that give rise to WGD and its effects in both sub-organismal and organismal systems will likely reveal additional commonalities across systems and processes.
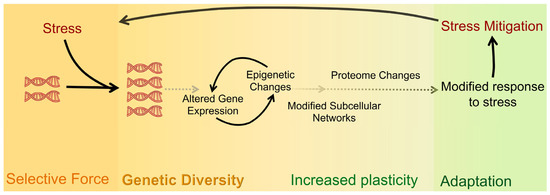
Figure 2. Shared causes and downstream effects of WGD across biological settings. Many forms of stress can lead an organism to undergo WGD. Whether as a response to the stress itself or as a way to adapt to the changing environment and/or different selective pressures, gene and epigenome changes lead to an altered proteome that supports an adaptive response to the initial stress exposure.
Polyploidy is often confused with aneuploidy, in part because, in humans, both phenomena are hallmarks of cancer [1][39][40][52]. Observations from numerous studies identify WGD in about 30% of human cancers [53][54] and as a precursor for many malignancies in cancer evolution [39][55][56][57][58][59]. Although WGD does not seem to require a preexisting cancer driver mutation; it most often emerges after oncogenic mutations, such as lesions in the TP53 genes [53][56]. WGD promotes diversification of copy number alterations (CNAs) and is permissive for other chromosomal aberrations and genome instability associated with poor cancer patient prognosis [39][58][59]. Interestingly, a recent study revealed a mechanism by which WGD enhances cancer-promoting alterations in DNA organization and gene expression [58]. Induction of WGD can reduce the levels of proteins involved in chromatin packaging, resulting in reorganization of the 3D DNA compartments and domains. Over time, loss of this domain structure predisposes the cell to additional cancer-promoting alterations and expression of oncogenic genes [58]. Polyploidization can also be protective, as in the liver where it both provides the basis for genetic adaptation to hepatotoxic stress and also protects from malignant transformations [21][60].
Organismal WGD is common in extant plants [7][32][61][62][63] and is also observed in protists [64], fungi [51][65][66], bacteria, and archaebacteria [67][68] and in several animal clades [2][12][35][36][40][41][69][70]. In both prokaryotes and eukaryotes, the effects of WGD are comparable with increased resistance to UV irradiation, cell size, adaptability to environmental changes, and evolvability [28][71]. Evidence for ancestral WGD events is found in the genomes of organisms of most clades, including vertebrates [36][72][73]. Polyploid metazoans arise due to errors during meiosis that result in the formation of diploid gametes that when fertilized give rise to polyploid organisms. Fertilization between diploid (2n) and haploid (n) gametes results in a triploid (3n) organism; fertilization between two diploid (2n) gametes results in a tetraploid (4n) [33][35]. When the diploid gametes of two related species fuse, the resulting organism is said to be an allopolyploid, whereas when diploid gametes from the same species fuse the result is said to be an autopolyploid [73]. Polyploidization from the cell to the organismal level most often results in lethality [32][74]. For instance, human triploid and tetraploid embryos die during gestation or soon after delivery, but triploid and tetraploid mosaic children may survive after birth [75][76]. This lethality may be caused by errors partitioning the additional chromosomes during cell division, by WGD-driven increases in cell size that interfere with tissues or organ scaling, and by epigenetic changes and gene expression changes that affect dose-dependent processes and complexes [73]. The latter may be especially true in human triploids and tetraploids which may have difficulty equalizing expression between the silenced X chromosomes and the extra sets of autosomes since variable numbers of Barr bodies may be found in these cells [75]. A newly formed polyploid may very occasionally overcome these obstacles, e.g., by establishing balanced genome expression. Then, if polyploidy provides a competitive advantage—as in a changing environment—polyploid lineages may outgrow their diploid counterparts and become established [32][41][73]. The established polyploid population will continue to evolve and then may “diploidize” to become a new species that has an adaptive advantage compared to the parental diploid species [32].
3. Laboratory Multicellular Organism Models for WGD
A major impediment to studying WGD and its impacts on the physiology and function of cells, tissues, and organs is the scarcity of laboratory models that permit comparison between isogenic organisms with differing ploidy [33]. Many laboratory models are sterile or embryonic lethal when polyploid [77][78][79]. Generation of autopolyploid laboratory model organisms that are not sterile (e.g., plants) frequently involves the use of chemicals such as colchicine, known to cause aneuploidy and genetic mutations. These polyploids, therefore, may not be truly isogenic with the parental diploid strains they were derived from [51][73]. The recently developed method for generating tetraploids in Caenorhabditis utilizes transient knock-down of a meiosis-specific cohesin (i.e., rec-8) by RNA interference to produce diploid gametes [80]. Therefore, the derived tetraploids are unlikely to differ significantly from the diploid strains from which they were derived (see Figure 3A for an overview of the method) [80].
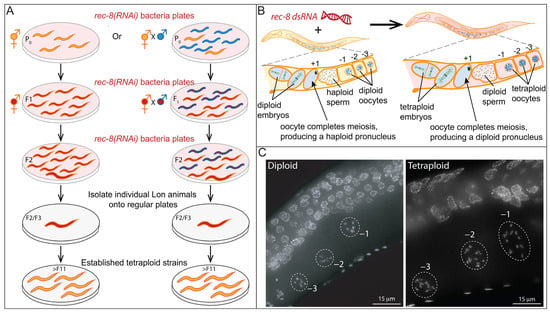
Figure 3. Method for rapid isolation of C. elegans tetraploids. (A). Diagram of the published method for generating tetraploid strains from diploids. This protocol relies on the transient knockdown of the meiosis-specific cohesin rec-8 which results in the production of diploid instead of haploid gametes. Thus, upon fertilization, tetraploid progeny (which are longer and bigger) are produced. The method works both during the selfing of the hermaphrodites (left) and during outcrossing (right). Crossing allows for the generation of tetraploids with two different genetic backgrounds. After two to three generations of exposure to rec-8 dsRNA, the larger hermaphrodites are individually plated on normal growth media. Strains that consistently produce large (Lon) hermaphrodites are then screened to confirm that these strains are tetraploids. (B). Diploid hermaphrodites (left) produce haploid gametes. Haploid sperm are stored in the spermatheca. Oocytes prior to the meiotic divisions have six pairs of connected homologous chromosomes that can be visualized by DAPI in the diakinesis-stage oocytes (Figure 3C, below) just adjacent to the spermatheca (−1, −2, and −3 oocytes based on position). The oocyte moves through the spermatheca and is fertilized and completes the meiotic divisions to produce a haploid pronucleus. The tetraploid worms derived from exposure to rec-8 dsRNA, (right) produce diploid gametes with 12 DAPI bodies. (C) Examples of fixed hermaphrodites stained with DAPI to screen for ploidy by counting chromosome pairs in the −1, −2, and −3 diakinesis oocytes.
References
- Davoli, T.; de Lange, T. The Causes and Consequences of Polyploidy in Normal Development and Cancer. Annu. Rev. Cell Dev. Biol. 2011, 27, 585–610.
- Øvrebø, J.I.; Edgar, B.A. Polyploidy in tissue homeostasis and regeneration. Development 2018, 145, dev156034.
- Donne, R.; Saroul-Aïnama, M.; Cordier, P.; Celton-Morizur, S.; Desdouets, C. Polyploidy in liver development, homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 391–405.
- Bailey, E.C.; Kobielski, S.; Park, J.; Losick, V.P. Polyploidy in Tissue Repair and Regeneration. Cold Spring Harb. Perspect. Biol. 2021, 13, a040881.
- Anatskaya, O.V.; Vinogradov, A.E. Polyploidy as a Fundamental Phenomenon in Evolution, Development, Adaptation and Diseases. Int. J. Mol. Sci. 2022, 23, 3542.
- Berman, J. Ploidy plasticity: A rapid and reversible strategy for adaptation to stress. FEMS Yeast Res. 2016, 16, fow020.
- Wood, T.E.; Takebayashi, N.; Barker, M.S.; Mayrose, I.; Greenspoon, P.B.; Rieseberg, L.H. The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 2009, 106, 13875–13879.
- Akagi, T.; Jung, K.; Masuda, K.; Shimizu, K.K. Polyploidy before and after domestication of crop species. Curr. Opin. Plant Biol. 2022, 69, 102255.
- Quinton, R.J.; DiDomizio, A.; Vittoria, M.A.; Kotýnková, K.; Ticas, C.J.; Patel, S.; Koga, Y.; Vakhshoorzadeh, J.; Hermance, N.; Kuroda, T.S.; et al. Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature 2021, 590, 492–497.
- Anatskaya, O.V.; Vinogradov, A.E. Polyploidy and Myc Proto-Oncogenes Promote Stress Adaptation via Epigenetic Plasticity and Gene Regulatory Network Rewiring. Int. J. Mol. Sci. 2022, 23, 9691.
- Orr-Weaver, T.L. When bigger is better: The role of polyploidy in organogenesis. Trends Genet. 2015, 31, 307–315.
- Frawley, L.E.; Orr-Weaver, T.L. Polyploidy. Curr. Biol. 2015, 25, R353–R358.
- Chen, H.-Z.; Ouseph, M.M.; Li, J.; Pécot, T.; Chokshi, V.; Kent, L.; Bae, S.; Byrne, M.; Duran, C.; Comstock, G.; et al. Canonical and atypical E2Fs regulate the mammalian endocycle. Nature 2012, 14, 1192–1202.
- Rios, A.C.; Fu, N.Y.; Jamieson, P.R.; Pal, B.; Whitehead, L.; Nicholas, K.R.; Lindeman, G.J.; Visvader, J.E. Essential role for a novel population of binucleated mammary epithelial cells in lactation. Nat. Commun. 2016, 7, 11400.
- Pandit, S.K.; Westendorp, B.; Nantasanti, S.; van Liere, E.; Tooten, P.C.J.; Cornelissen, P.W.A.; Toussaint, M.J.M.; Lamers, W.H.; de Bruin, A. E2F8 is essential for polyploidization in mammalian cells. Nature 2012, 14, 1181–1191.
- Senyo, S.E.; Steinhauser, M.L.; Pizzimenti, C.L.; Yang, V.K.; Cai, L.; Wang, M.; Wu, T.-D.; Guerquin-Kern, J.-L.; Lechene, C.P.; Lee, R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2013, 493, 433–436.
- Zanet, J.; Freije, A.; Ruiz, M.; Coulon, V.; Sanz, J.R.; Chiesa, J.; Gandarillas, A. A Mitosis Block Links Active Cell Cycle with Human Epidermal Differentiation and Results in Endoreplication. PLoS ONE 2010, 5, e15701.
- Mattia, G.; Vulcano, F.; Milazzo, L.; Barca, A.; Macioce, G.; Giampaolo, A.; Hassan, H.J. Different ploidy levels of megakaryocytes generated from peripheral or cord blood CD34+ cells are correlated with different levels of platelet release. Blood 2002, 99, 888–897.
- Losick, V.P. Wound-Induced Polyploidy Is Required for Tissue Repair. Adv. Wound Care 2016, 5, 271–278.
- Zimmet, J.; Ravid, K. Polyploidy: Occurrence in nature, mechanisms, and significance for the megakaryocyte-platelet system. Exp. Hematol. 2000, 28, 3–16.
- Sladky, V.C.; Eichin, F.; Reiberger, T.; Villunger, A. Polyploidy control in hepatic health and disease. J. Hepatol. 2021, 75, 1177–1191.
- Mazzi, S.; Lordier, L.; Debili, N.; Raslova, H.; Vainchenker, W. Megakaryocyte and polyploidization. Exp. Hematol. 2018, 57, 1–13.
- Anatskaya, O.V.; Sidorenko, N.V.; Vinogradov, A.E.; Beyer, T.V. Impact of neonatal cryptosporidial gastroenteritis on epigenetic programming of rat hepatocytes. Cell Biol. Int. 2007, 31, 420–427.
- Anatskaya, O.V.; Matveev, I.V.; Sidorenko, N.V.; Kharchenko, M.V.; Kropotov, A.V.; Vinogradov, A.E. Changes in the heart of neonatal rats after cryptosporidial gastroenteritis of different degrees of severity. J. Evol. Biochem. Physiol. 2013, 49, 509–518.
- Lazzeri, E.; Angelotti, M.L.; Conte, C.; Anders, H.-J.; Romagnani, P. Surviving Acute Organ Failure: Cell Polyploidization and Progenitor Proliferation. Trends Mol. Med. 2019, 25, 366–381.
- Anatskaya, O.V.; Vinogradov, A.E. Whole-Genome Duplications in Evolution, Ontogeny, and Pathology: Complexity and Emergency Reserves. Mol. Biol. 2021, 55, 813–827.
- Edgar, B.A.; Zielke, N.; Gutierrez, C. Endocycles: A recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 2014, 15, 197–210.
- Ohbayashi, R.; Nakamachi, A.; Hatakeyama, T.S.; Watanabe, S.; Kanesaki, Y.; Chibazakura, T.; Yoshikawa, H.; Miyagishima, S.-Y. Coordination of Polyploid Chromosome Replication with Cell Size and Growth in a Cyanobacterium. mBio 2019, 10, e00510-19.
- Chen, A.H.; Afonso, B.; Silver, P.A.; Savage, D.F. Spatial and Temporal Organization of Chromosome Duplication and Segregation in the Cyanobacterium Synechococcus elongatus PCC 7942. PLoS ONE 2012, 7, e47837.
- Watanabe, S.; Ohbayashi, R.; Shiwa, Y.; Noda, A.; Kanesaki, Y.; Chibazakura, T.; Yoshikawa, H. Light-dependent and asynchronous replication of cyanobacterial multi-copy chromosomes. Mol. Microbiol. 2012, 83, 856–865.
- Schoenfelder, K.P.; Fox, D.T. The expanding implications of polyploidy. J. Cell Biol. 2015, 209, 485–491.
- Bomblies, K. When everything changes at once: Finding a new normal after genome duplication. Proc. R. Soc. B Boil. Sci. 2020, 287, 20202154.
- Doyle, J.J.; Coate, J.E. Polyploidy, the Nucleotype, and Novelty: The Impact of Genome Doubling on the Biology of the Cell. Int. J. Plant Sci. 2019, 180, 1–52.
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Mol. Plant 2018, 11, 414–428.
- Fox, D.T.; Soltis, D.E.; Soltis, P.S.; Ashman, T.-L.; Van de Peer, Y. Polyploidy: A Biological Force from Cells to Ecosystems. Trends Cell Biol. 2020, 30, 688–694.
- Maciver, S.K. Ancestral Eukaryotes Reproduced Asexually, Facilitated by Polyploidy: A Hypothesis. Bioessays 2019, 41, e1900152.
- Nguyen, H.G.; Makitalo, M.; Yang, D.; Chinnappan, D.; St.Hilaire, C.; Ravid, K. Deregulated Aurora-B induced tetraploidy promotes tumorigenesis. FASEB J. 2009, 23, 2741–2748.
- van Rijnberk, L.M.; Barrull-Mascaró, R.; van der Palen, R.L.; Schild, E.S.; Korswagen, H.C.; Galli, M. Endomitosis controls tissue-specific gene expression during development. PLoS Biol. 2022, 20, e3001597.
- Storchova, Z.; Pellman, D. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 2004, 5, 45–54.
- Zhang, J.; Qiao, Q.; Xu, H.; Zhou, R.; Liu, X. Human cell polyploidization: The good and the evil. Semin. Cancer Biol. 2022, 81, 54–63.
- Otto, S.P. The Evolutionary Consequences of Polyploidy. Cell 2007, 131, 452–462.
- Lazzeri, E.; Angelotti, M.L.; Peired, A.; Conte, C.; Marschner, J.A.; Maggi, L.; Mazzinghi, B.; Lombardi, D.; Melica, M.E.; Nardi, S.; et al. Endocycle-related tubular cell hypertrophy and progenitor proliferation recover renal function after acute kidney injury. Nat. Commun. 2018, 9, 1344.
- Losick, V.P.; Fox, D.T.; Spradling, A.C. Polyploidization and Cell Fusion Contribute to Wound Healing in the Adult Drosophila Epithelium. Curr. Biol. 2013, 23, 2224–2232.
- Li, T.-N.; Wu, Y.-J.; Tsai, H.-W.; Sun, C.-P.; Wu, H.-L.; Pei, Y.-N.; Lu, K.-Y.; Yen, T.T.-C.; Chang, C.-W.; Chan, H.-L.; et al. Intrahepatic hepatitis B virus large surface antigen induces hepatocyte hyperploidy via failure of cytokinesis. J. Pathol. 2018, 245, 502–513.
- David, K.T.; Halanych, K.M. Spatial proximity between polyploids across South American frog genera. J. Biogeogr. 2021, 48, 991–1000.
- Glennon, K.L.; Ritchie, M.E.; Segraves, K.A. Evidence for shared broad-scale climatic niches of diploid and polyploid plants. Ecol. Lett. 2014, 17, 574–582.
- Arnold, B.; Kim, S.-T.; Bomblies, K. Single Geographic Origin of a Widespread Autotetraploid Arabidopsis arenosa Lineage Followed by Interploidy Admixture. Mol. Biol. Evol. 2015, 32, 1382–1395.
- Molina-Henao, Y.F.; Hopkins, R. Autopolyploid lineage shows climatic niche expansion but not divergence in Arabidopsis arenosa. Am. J. Bot. 2019, 106, 61–70.
- Ramsey, J. Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. USA 2011, 108, 7096–7101.
- Martin, S.L.; Husband, B.C. Adaptation of diploid and tetraploid chamerion angustifolium to eleva-tion but not local environment: Adaptation of Chamerion angustifolium ploidies. Evolution 2013, 67, 1780–1791.
- Selmecki, A.M.; Maruvka, Y.E.; Richmond, P.A.; Guillet, M.; Shoresh, N.; Sorenson, A.L.; De, S.; Kishony, R.; Michor, F.; Dowell, R.; et al. Polyploidy can drive rapid adaptation in yeast. Nature 2015, 519, 349–352.
- Van De Peer, Y.; Mizrachi, Y.V.D.P.E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424.
- Bielski, C.M.; Zehir, A.; Penson, A.V.; Donoghue, M.T.A.; Chatila, W.; Armenia, J.; Chang, M.T.; Schram, A.M.; Jonsson, P.; Bandlamudi, C.; et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet. 2018, 50, 1189–1195.
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhang, C.Z.; Wala, J.; Mermel, C.H.; et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013, 45, 1134–1140.
- Sansregret, L.; Vanhaesebroeck, B.; Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 139–150.
- Fujiwara, T.; Bandi, M.; Nitta, M.; Ivanova, E.V.; Bronson, R.T.; Pellman, D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005, 437, 1043–1047.
- Matsumoto, T.; Wakefield, L.; Peters, A.; Peto, M.; Spellman, P.; Grompe, M. Proliferative polyploid cells give rise to tumors via ploidy reduction. Nat. Commun. 2021, 12, 646.
- Lambuta, R.A.; Nanni, L.; Liu, Y.; Diaz-Miyar, J.; Iyer, A.; Tavernari, D.; Katanayeva, N.; Ciriello, G.; Oricchio, E. Whole-genome doubling drives oncogenic loss of chromatin segregation. Nature 2023, 615, 925–933.
- Dewhurst, S.M.; McGranahan, N.; Burrell, R.A.; Rowan, A.J.; Grönroos, E.; Endesfelder, D.; Joshi, T.; Mouradov, D.; Gibbs, P.; Ward, R.L.; et al. Tolerance of Whole-Genome Doubling Propagates Chromosomal Instability and Accelerates Cancer Genome Evolution. Cancer Discov. 2014, 4, 175–185.
- Lin, Y.-H.; Zhang, S.; Zhu, M.; Lu, T.; Chen, K.; Wen, Z.; Wang, S.; Xiao, G.; Luo, D.; Jia, Y.; et al. Mice with Increased Numbers of Polyploid Hepatocytes Maintain Regenerative Capacity But Develop Fewer Hepatocellular Carcinomas Following Chronic Liver Injury. Gastroenterology 2020, 158, 1698–1712.e14.
- Adams, K.L.; Wendel, J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005, 8, 135–141.
- Cheng, A.; Hanafiah, N.M.; Harikrishna, J.A.; Eem, L.P.; Baisakh, N.; Mispan, M.S. A Reappraisal of Polyploidy Events in Grasses (Poaceae) in a Rapidly Changing World. Biology 2022, 11, 636.
- Park, C.-W.; Bhandari, G.S.; Won, H.; Park, J.H.; Park, D.S. Polyploidy and introgression in invasive giant knotweed (Fallopia sachalinensis) during the colonization of remote volcanic islands. Sci. Rep. 2018, 8, 16021.
- Demin, S.Y.; Berdieva, M.A.; Goodkov, A.V. Cyclic Polyploidy in Obligate Agamic Amoebae. Cell Tissue Biol. 2019, 13, 242–246.
- Storchová, Z.; Breneman, A.; Cande, J.; Dunn, J.; Burbank, K.; O’Toole, E.; Pellman, D. Genome-wide genetic analysis of polyploidy in yeast. Nature 2006, 443, 541–547.
- Albertin, W.; Marullo, P. Polyploidy in fungi: Evolution after whole-genome duplication. Proc. R. Soc. B Boil. Sci. 2012, 279, 2497–2509.
- Soppa, J. Polyploidy in Archaea and Bacteria: About Desiccation Resistance, Giant Cell Size, Long-Term Survival, Enforcement by a Eukaryotic Host and Additional Aspects. Microb. Physiol. 2015, 24, 409–419.
- Markov, A.V.; Kaznacheev, I.S. Evolutionary consequences of polyploidy in prokaryotes and the origin of mitosis and meiosis. Biol. Direct 2016, 11, 28.
- Wertheim, B.; Beukeboom, L.; van de Zande, L. Polyploidy in Animals: Effects of Gene Expression on Sex Determination, Evolution and Ecology. Cytogenet. Genome Res. 2013, 140, 256–269.
- Mable, B.K.; Alexandrou, M.A.; Taylor, M.I. Genome duplication in amphibians and fish: An extended synthesis. J. Zool. 2011, 284, 151–182.
- Mendell, J.E.; Clements, K.D.; Choat, J.H.; Angert, E.R. Extreme polyploidy in a large bacterium. Proc. Natl. Acad. Sci. USA 2008, 105, 6730–6734.
- Sacerdot, C.; Louis, A.; Bon, C.; Berthelot, C.; Crollius, H.R. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol. 2018, 19, 166.
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846.
- Monnahan, P.; Kolář, F.; Baduel, P.; Sailer, C.; Koch, J.; Horvath, R.; Laenen, B.; Schmickl, R.; Paajanen, P.; Šrámková, G.; et al. Pervasive population genomic consequences of genome duplication in Arabidopsis arenosa. Nat. Ecol. Evol. 2019, 3, 457–468.
- Kelly, T.E.; Rary, J.M. Mosaic tetraploidy in a two-year-old female. Clin. Genet. 1974, 6, 221–224.
- Stefanova, I.; Jenderny, J.; Kaminsky, E.; Mannhardt, A.; Meinecke, P.; Grozdanova, L.; Gillessen-Kaesbach, G. Mosaic and complete tetraploidy in live-born infants: Two new patients and review of the literature. Clin. Dysmorphol. 2010, 19, 123–127.
- Yamazaki, W.; Takahashi, M.; Kawahara, M. Restricted development of mouse triploid fetuses with disorganized expression of imprinted genes. Zygote 2015, 23, 874–884.
- Menon, T.; Nair, S. Experimental Manipulation of Ploidy in Zebrafish Embryos and Its Application in Genetic Screens. Vertebr. Embryog. Embryol. Cell. Genet. Methods 2019, 1920, 111–128.
- Eakin, G.; Hadjantonakis, A.-K.; Papaioannou, V.; Behringer, R.R. Developmental potential and behavior of tetraploid cells in the mouse embryo. Dev. Biol. 2005, 288, 150–159.
- Clarke, E.K.; Gomez, K.A.R.; Mustachi, Z.; Murph, M.C.; Schvarzstein, M. Manipulation of Ploidy in Caenorhabditis elegans. J. Vis. Exp. 2018, 133, 57296.
More
Information
Subjects:
Developmental Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
737
Revisions:
2 times
(View History)
Update Date:
30 Jun 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




