
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | López de Felipe Félix | -- | 1900 | 2023-06-28 13:15:09 | | | |
| 2 | Catherine Yang | Meta information modification | 1900 | 2023-06-29 04:28:42 | | |
Video Upload Options
Hydroxycinnamic acids (HCAs) are phenolic compounds produced by the secondary metabolism of edible plants and are the most abundant phenolic acids in our diet. The antimicrobial capacity of HCAs is an important function attributed to these phenolic acids in the defense of plants against microbiological threats, and bacteria have developed diverse mechanisms to counter the antimicrobial stress imposed by these compounds, including their metabolism into different microbial derivatives. The metabolism of HCAs has been intensively studied in Lactobacillus spp., as the metabolic transformation of HCAs by these bacteria contributes to the biological activity of these acids in plant and human habitats or to improve the nutritional quality of fermented foods.
1. Introduction
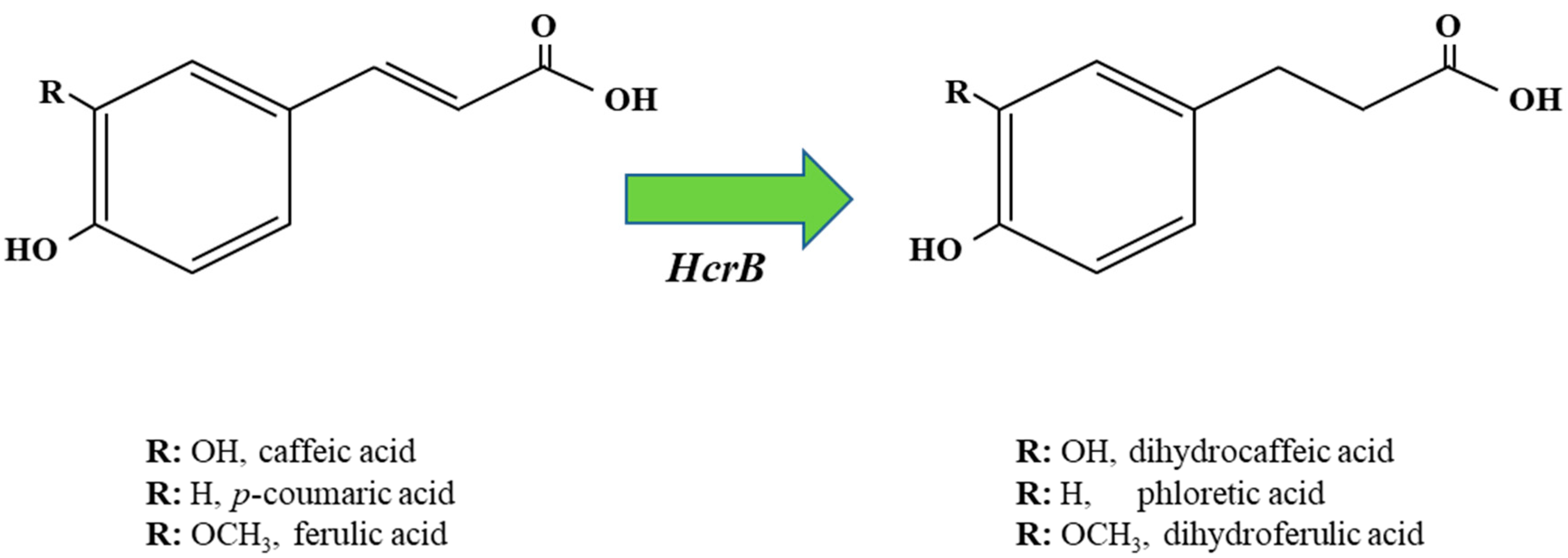
2. Variability in the HCA Reduction and Decarboxylation Metabolisms across Lactobacillus spp.
3. HrcAB Reductase: Open Questions
References
- Gaur, G.; Oh, J.-H.; Filannino, P.; Gobbetti, M.; van Pijkeren, J.-P.; Gänzle, M.G. Genetic Determinants of Hydroxycinnamic Acid Metabolism in Heterofermentative Lactobacilli. Appl. Environ. Microbiol. 2020, 86, e02461-19.
- Rodríguez, H.; Landete, J.M.; Rivas, B.d.L.; Muñoz, R. Metabolism of food phenolic acids by Lactobacillus plantarum CECT 748T. Food Chem. 2008, 107, 1393–1398.
- Reverón, I.; de las Rivas, B.; Muñoz, R.; López de Felipe, F. Genome-wide transcriptomic responses of a human isolate of Lactobacillus plantarum exposed to p-coumaric acid stress. Mol. Nutr. Food Res. 2012, 56, 1848–1859.
- Filannino, P.; Di Cagno, R.; Crecchio, C.; De Virgilio, C.; De Angelis, M.; Gobbetti, M. Transcriptional reprogramming and phenotypic switching associated with the adaptation of Lactobacillus plantarum C2 to plant niches. Sci. Rep. 2016, 6, 1–16.
- Wen, L.; Wrolstad, R. Phenolic Composition of Authentic Pineapple Juice. J. Food Sci. 2002, 67, 155–161.
- Santamaría, L.; Reverón, I.; de Felipe, F.L.; Rivas, B.D.L.; Muñoz, R. Unravelling the Reduction Pathway as an Alternative Metabolic Route to Hydroxycinnamate Decarboxylation in Lactobacillus plantarum. Appl. Environ. Microbiol. 2018, 84, e01123-18.
- Hertzberger, R.; Arents, J.; Dekker, H.L.; Pridmore, R.D.; Gysler, C.; Kleerebezem, M.; de Mattos, M.J.T. H2O2 Production in Species of the Lactobacillus acidophilus Group: A Central Role for a Novel NADH-Dependent Flavin Reductase. Appl. Environ. Microbiol. 2014, 80, 2229–2239.
- Valladares, R.B.; Graves, C.; Wright, K.; Gardner, C.L.; Lorca, G.L.; Gonzalez, C.F. H2O2 production rate in Lactobacillus johnsonii is modulated via the interplay of a heterodimeric flavin oxidoreductase with a soluble 28 Kd PAS domain containing protein. Front. Microbiol. 2015, 6, 716.
- Gutierrez-Zetina, S.M.; González-Manzano, S.; Ayuda-Durán, B.; Santos-Buelga, C.; González-Paramás, A.M. Caffeic and Dihydrocaffeic Acids Promote Longevity and Increase Stress Resistance in Caenorhabditis elegans by Modulating Expression of Stress-Related Genes. Molecules 2021, 26, 1517.
- Silva, F.A.M.; Borges, F.; Guimarães, C.; Lima, J.L.F.C.; Matos, C.; Reis, S. Phenolic Acids and Derivatives: Studies on the Relationship among Structure, Radical Scavenging Activity, and Physicochemical Parameters. J. Agric. Food Chem. 2000, 48, 2122–2126.
- Esteves, M.; Siquet, C.; Gaspar, A.; Rio, V.; Sousa, J.B.; Reis, S.; Marques, M.P.M.; Borges, F. Antioxidant Versus Cytotoxic Properties of Hydroxycinnamic Acid Derivatives—A New Paradigm in Phenolic Research. Arch. Pharm. 2008, 341, 164–173.
- Segura-Aguilar, J.; Lind, C. On the mechanism of the Mn3+-induced neurotoxicity of dopamine: Prevention of quinone-derived oxygen toxicity by DT diaphorase and superoxide dismutase. Chem. Interact. 1989, 72, 309–324.
- Santamaría, L.; Reverón, I.; Plaza-Vinuesa, L.; Oliveros, J.C.; Rivas, B.D.L.; Muñoz, R.; De Felipe, F.L. Oleuropein Transcriptionally Primes Lactobacillus plantarum to Interact With Plant Hosts. Front. Microbiol. 2019, 10, 2177.
- Reverón, I.; Rivas, B.D.L.; Matesanz, R.; Muñoz, R.; de Felipe, F.L. Molecular adaptation of Lactobacillus plantarum WCFS1 to gallic acid revealed by genome-scale transcriptomic signature and physiological analysis. Microb. Cell Factories 2015, 14, 160.
- Barthelmebs, L.; Divies, C.; Cavin, J.F. Knockout of the p-coumarate decarboxylase gene from Lactobacillus plantarum reveals the existence of two other inducible enzymatic activities involved in phenolic acid metabolism. Appl. Environ. Microbiol. 2000, 66, 3368–3375.
- Sun, J.; Lin, Y.; Shen, X.; Jain, R.; Sun, X.; Yuan, Q.; Yan, Y. Aerobic biosynthesis of hydrocinnamic acids in Escherichia coli with a strictly oxygen-sensitive enoate reductase. Metab. Eng. 2016, 35, 75–82.
- Aguirre, J.D.; Culotta, V.C. Battles with Iron: Manganese in Oxidative Stress Protection. J. Biol. Chem. 2012, 287, 13541–13548.
- Dukan, S.; Nyström, T. Oxidative Stress Defense and Deterioration of Growth-arrested Escherichia coli Cells. J. Biol. Chem. 1999, 274, 26027–26032.
- De Felipe, F.L.; Hugenholtz, J. Purification and characterisation of the water forming NADH-oxidase from Lactococcus lactis. Int. Dairy J. 2001, 11, 37–44.
- Zhang, Y.-W.; Tiwari, M.K.; Gao, H.; Dhiman, S.S.; Jeya, M.; Lee, J.-K. Cloning and characterization of a thermostable H2O-forming NADH oxidase from Lactobacillus rhamnosus. Enzym. Microb. Technol. 2012, 50, 255–262.
- Filannino, P.; Gobbetti, M.; De Angelis, M.; Di Cagno, R. Hydroxycinnamic Acids Used as External Acceptors of Electrons: An Energetic Advantage for Strictly Heterofermentative Lactic Acid Bacteria. Appl. Environ. Microbiol. 2014, 80, 7574–7582.
- Filannino, P.; Di Cagno, R.; Addante, R.; Pontonio, E.; Gobbetti, M. Metabolism of Fructophilic Lactic Acid Bacteria Isolated from the Apis mellifera L. Bee Gut: Phenolic Acids as External Electron Acceptors. Appl. Environ. Microbiol. 2016, 82, 6899–6911.




