
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Rashed Abualia | -- | 1873 | 2023-06-27 15:41:11 | | | |
| 2 | Wendy Huang | Meta information modification | 1873 | 2023-06-28 07:50:46 | | | | |
| 3 | Wendy Huang | + 36 word(s) | 1909 | 2023-07-10 05:38:12 | | |
Video Upload Options
As a major component of vital macromolecules such as nucleic acids, amino acids, and chlorophyll, nitrogen is an essential macronutrient for plants. Although nitrogen is one of the most abundant elements in nature, accounting for about 70% of atmospheric gasses, its availability for plant uptake in the soil varies temporally and spatially. Therefore, modern agriculture relies heavily on nitrogen fertilization to maximize crop quality and yield. Auxins are a group of naturally occurring molecules derived from tryptophan, with indole-3-acetic acid (IAA) being the major form of auxin. The biosynthesis of IAA is defined by a two-step metabolic pathway, in which the TAA family of aminotransferases converts tryptophan (Trp) to indole-3-pyruvate (IPA), followed by a YUC flavin monooxygenases-mediated conversion of IPA to IAA. Auxin has extensive regulatory functions in plant development.
1. Introduction
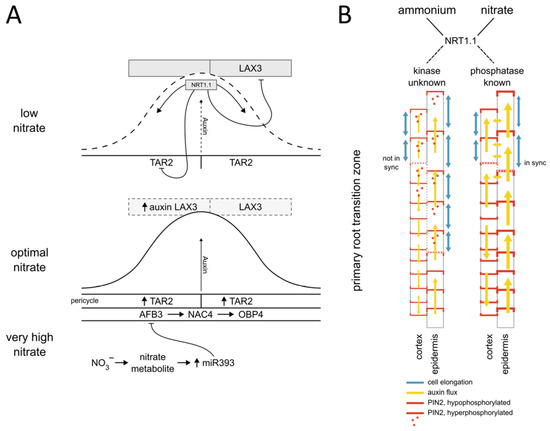
2. The Role of Auxin in Nitrate-Regulated Plant Growth and Development
References
- von Wirén, N.; Gazzarrini, S.; Gojon, A.; Frommer, W.B. The molecular physiology of ammonium uptake and retrieval. Curr. Opin. Plant Biol. 2000, 3, 254–261.
- Crawford, N.M.; Glass, A.D.M. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998, 3, 389–395.
- Näsholm, T.; Kielland, K.; Ganeteg, U. Uptake of organic nitrogen by plants. New Phytol. 2009, 182, 31–48.
- Hänsch, R.; Mendel, R.R. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 2009, 12, 259–266.
- Ötvös, K.; Marconi, M.; Vega, A.; O’Brien, J.; Johnson, A.; Abualia, R.; Antonielli, L.; Montesinos, J.C.; Zhang, Y.; Tan, S.; et al. Modulation of plant root growth by nitrogen source-defined regulation of polar auxin transport. EMBO J. 2021, 40, e106862.
- Alvarez, J.M.; Riveras, E.; Vidal, E.A.; Gras, D.E.; Contreras-López, O.; Tamayo, K.P.; Aceituno, F.; Gómez, I.; Ruffel, S.; Lejay, L.; et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. Cell Mol. Biol. 2014, 80, 1–13.
- Vidal, E.A.; Araus, V.; Lu, C.; Parry, G.; Green, P.J.; Coruzzi, G.M.; Gutiérrez, R.A. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 4477–4482.
- Vidal, E.A.; Álvarez, J.M.; Gutiérrez, R.A. Nitrate regulation of AFB3 and NAC4 gene expression in Arabidopsis roots depends on NRT1.1 nitrate transport function. Plant Signal. Behav. 2014, 9, e28501.
- Zhang, H.; Forde, B.G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 1998, 279, 407–409.
- Tian, Q.-Y.; Sun, P.; Zhang, W.-H. Ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana. New Phytol. 2009, 184, 918–931.
- Li, Q.; Li, B.-H.; Kronzucker, H.J.; Shi, W.-M. Root growth inhibition by NH(4)(+) in Arabidopsis is mediated by the root tip and is linked to NH(4)(+) efflux and GMPase activity. Plant Cell Environ. 2010, 33, 1529–1542.
- Walch-Liu, P.; Ivanov, I.I.; Filleur, S.; Gan, Y.; Remans, T.; Forde, B.G. Nitrogen Regulation of Root Branching. Ann. Bot. 2006, 97, 875–881.
- Takei, K.; Ueda, N.; Aoki, K.; Kuromori, T.; Hirayama, T.; Shinozaki, K.; Yamaya, T.; Sakakibara, H. AtIPT3 is a Key Determinant of Nitrate-Dependent Cytokinin Biosynthesis in Arabidopsis. Plant Cell Physiol. 2004, 45, 1053–1062.
- Poitout, A.; Crabos, A.; Petřík, I.; Novák, O.; Krouk, G.; Lacombe, B.; Ruffel, S. Responses to Systemic Nitrogen Signaling in Arabidopsis Roots Involve trans-Zeatin in Shoots. Plant Cell 2018, 30, 1243–1257.
- Abualia, R.; Ötvös, K.; Novák, O.; Bouguyon, E.; Domanegg, K.; Krapp, A.; Nacry, P.; Gojon, A.; Lacombe, B.; Benková, E. Molecular framework integrating nitrate sensing in root and auxin-guided shoot adaptive responses. Proc. Natl. Acad. Sci. USA 2022, 119, e2122460119.
- Maeda, Y.; Konishi, M.; Kiba, T.; Sakuraba, Y.; Sawaki, N.; Kurai, T.; Ueda, Y.; Sakakibara, H.; Yanagisawa, S. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 2018, 9, 1376.
- Varala, K.; Marshall-Colón, A.; Cirrone, J.; Brooks, M.D.; Pasquino, A.V.; Léran, S.; Mittal, S.; Rock, T.M.; Edwards, M.B.; Kim, G.J.; et al. Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc. Natl. Acad. Sci. USA 2018, 115, 6494–6499.
- Alvarez, J.M.; Schinke, A.-L.; Brooks, M.D.; Pasquino, A.; Leonelli, L.; Varala, K.; Safi, A.; Krouk, G.; Krapp, A.; Coruzzi, G.M. Transient genome-wide interactions of the master transcription factor NLP7 initiate a rapid nitrogen-response cascade. Nat. Commun. 2020, 11, 1157.
- Vega, A.; Fredes, I.; O’Brien, J.; Shen, Z.; Ötvös, K.; Abualia, R.; Benkova, E.; Briggs, S.P.; Gutiérrez, R.A. Nitrate triggered phosphoproteome changes and a PIN2 phosphosite modulating root system architecture. EMBO Rep. 2021, 22, e51813.
- Liu, K.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun Chung, H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 2017, 545, 311–316.
- Gutiérrez, R.A.; Lejay, L.V.; Dean, A.; Chiaromonte, F.; Shasha, D.E.; Coruzzi, G.M. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 2007, 8, R7.
- Castaings, L.; Camargo, A.; Pocholle, D.; Gaudon, V.; Texier, Y.; Boutet-Mercey, S.; Taconnat, L.; Renou, J.-P.; Daniel-Vedele, F.; Fernandez, E.; et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009, 57, 426–435.
- Marchive, C.; Roudier, F.; Castaings, L.; Bréhaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013, 4, 1713.
- Liu, K.-H.; Liu, M.; Lin, Z.; Wang, Z.-F.; Chen, B.; Liu, C.; Guo, A.; Konishi, M.; Yanagisawa, S.; Wagner, G.; et al. NIN-like protein 7 transcription factor is a plant nitrate sensor. Science 2022, 377, 1419–1425.
- Liu, K.-H.; Diener, A.; Lin, Z.; Liu, C.; Sheen, J. Primary nitrate responses mediated by calcium signalling and diverse protein phosphorylation. J. Exp. Bot. 2020, 71, 4428–4441.
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 2010, 18, 927–937.
- Maghiaoui, A.; Bouguyon, E.; Cuesta, C.; Perrine-Walker, F.; Alcon, C.; Krouk, G.; Benková, E.; Nacry, P.; Gojon, A.; Bach, L. The Arabidopsis NRT1.1 transceptor coordinately controls auxin biosynthesis and transport to regulate root branching in response to nitrate. J. Exp. Bot. 2020, 71, 4480–4494.
- Mounier, E.; Pervent, M.; Ljung, K.; Gojon, A.; Nacry, P. Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ. 2014, 37, 162–174.
- Signora, L.; De Smet, I.; Foyer, C.H.; Zhang, H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. Cell Mol. Biol. 2001, 28, 655–662.
- Zhao, Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 2012, 5, 334–338.
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, Efflux-Dependent Auxin Gradients as a Common Module for Plant Organ Formation. Cell 2003, 115, 591–602.
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 2003, 426, 147–153.
- Reinhardt, D.; Pesce, E.-R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Traas, J.; Friml, J.; Kuhlemeier, C. Regulation of phyllotaxis by polar auxin transport. Nature 2003, 426, 255–260.
- Chapman, E.J.; Estelle, M. Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 2009, 43, 265–285.
- Rademacher, E.H.; Lokerse, A.S.; Schlereth, A.; Llavata-Peris, C.I.; Bayer, M.; Kientz, M.; Freire Rios, A.; Borst, J.W.; Lukowitz, W.; Jürgens, G.; et al. Different Auxin Response Machineries Control Distinct Cell Fates in the Early Plant Embryo. Dev. Cell 2012, 22, 211–222.
- Bargmann, B.O.R.; Vanneste, S.; Krouk, G.; Nawy, T.; Efroni, I.; Shani, E.; Choe, G.; Friml, J.; Bergmann, D.C.; Estelle, M.; et al. A map of cell type-specific auxin responses. Mol. Syst. Biol. 2013, 9, 688.
- Fendrych, M.; Akhmanova, M.; Merrin, J.; Glanc, M.; Hagihara, S.; Takahashi, K.; Uchida, N.; Torii, K.U.; Friml, J. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat. Plants 2018, 4, 453–459.
- Kubeš, M.; Napier, R. Non-canonical auxin signalling: Fast and curious. J. Exp. Bot. 2019, 70, 2609–2614.
- Lin, W.; Zhou, X.; Tang, W.; Takahashi, K.; Pan, X.; Dai, J.; Ren, H.; Zhu, X.; Pan, S.; Zheng, H.; et al. TMK-based cell-surface auxin signalling activates cell-wall acidification. Nature 2021, 599, 278–282.
- Li, L.; Verstraeten, I.; Roosjen, M.; Takahashi, K.; Rodriguez, L.; Merrin, J.; Chen, J.; Shabala, L.; Smet, W.; Ren, H.; et al. Antagonistic cell surface and intracellular auxin signalling regulate plasma membrane H+-fluxes for root growth. Nature 2021, 599, 273.
- Friml, J.; Gallei, M.; Gelová, Z.; Johnson, A.; Mazur, E.; Monzer, A.; Rodriguez, L.; Roosjen, M.; Verstraeten, I.; Živanović, B.D.; et al. ABP1–TMK auxin perception for global phosphorylation and auxin canalization. Nature 2022, 609, 575–581.
- Cambridge, A.P.; Morris, D.A. Transfer of exogenous auxin from the phloem to the polar auxin transport pathway in pea (Pisum sativum L.). Planta 1996, 199, 583–588.
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32.
- Abualia, R.; Benkova, E.; Lacombe, B. Transporters and Mechanisms of Hormone Transport in Arabidopsis. Adv. Bot. Res. 2018, 87, 342.
- Barbez, E.; Kubeš, M.; Rolčík, J.; Béziat, C.; Pěnčík, A.; Wang, B.; Rosquete, M.R.; Zhu, J.; Dobrev, P.I.; Lee, Y.; et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 2012, 485, 119–122.
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. Plant Cell 1991, 3, 677–684.
- Bennett, M.J.; Marchant, A.; Green, H.G.; May, S.T.; Ward, S.P.; Millner, P.A.; Walker, A.R.; Schulz, B.; Feldmann, K.A. Arabidopsis AUX1 Gene: A Permease-Like Regulator of Root Gravitropism. Science 1996, 273, 948–950.
- Ye, L.; Liu, L.; Xing, A.; Kang, D. Characterization of a dwarf mutant allele of Arabidopsis MDR-like ABC transporter AtPGP1 gene. Biochem. Biophys. Res. Commun. 2013, 441, 782–786.
- Avery, G.S.; Pottorf, L. Auxin and Nitrogen Relationships in Green Plants. Am. J. Bot. 1945, 32, 666–669.
- Caba, J.M.; Centeno, M.L.; Fernández, B.; Gresshoff, P.M.; Ligero, F. Inoculation and nitrate alter phytohormone levels in soybean roots: Differences between a supernodulating mutant and the wild type. Planta 2000, 211, 98–104.
- Tian, Q.; Chen, F.; Liu, J.; Zhang, F.; Mi, G. Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J. Plant Physiol. 2008, 165, 942–951.
- Meier, M.; Liu, Y.; Lay-Pruitt, K.S.; Takahashi, H.; von Wirén, N. Auxin-mediated root branching is determined by the form of available nitrogen. Nat. Plants 2020, 6, 1136–1145.
- Ma, W.; Li, J.; Qu, B.; He, X.; Zhao, X.; Li, B.; Fu, X.; Tong, Y. Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. Cell Mol. Biol. 2014, 78, 70–79.
- Bouguyon, E.; Brun, F.; Meynard, D.; Kubeš, M.; Pervent, M.; Leran, S.; Lacombe, B.; Krouk, G.; Guiderdoni, E.; Zažímalová, E.; et al. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 2015, 1, 15015.
- Porco, S.; Larrieu, A.; Du, Y.; Gaudinier, A.; Goh, T.; Swarup, K.; Swarup, R.; Kuempers, B.; Bishopp, A.; Lavenus, J.; et al. Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development 2016, 143, 3340–3349.
- Lay-Pruitt, K.S.; Takahashi, H. Integrating N signals and root growth: The role of nitrate transceptor NRT1.1 in auxin-mediated lateral root development. J. Exp. Bot. 2020, 71, 4365–4368.
- Lee, H.; Ganguly, A.; Baik, S.; Cho, H.-T. Calcium-dependent protein kinase 29 modulates PIN-FORMED polarity and Arabidopsis development via its own phosphorylation code. Plant Cell 2021, 33, 3513–3531.




