
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Modesto Redrejo Rodríguez | -- | 1405 | 2023-06-26 10:38:17 | | | |
| 2 | Conner Chen | + 8 word(s) | 1413 | 2023-06-29 07:16:40 | | |
Video Upload Options
In the same way that specialized DNA polymerases (DNAPs) replicate cellular and viral genomes, only a handful of dedicated proteins from various natural origins as well as engineered versions are appropriate for competent exponential amplification of whole genomes and metagenomes (WGA). Different applications have led to the development of diverse protocols, based on various DNAPs. Isothermal whole genomes and metagenomes (WGA) is currently widely used due to the high performance of Φ29 DNA polymerase, but polymerase chain reaction (PCR)-based methods are also available and can provide competent amplification of certain samples. Replication fidelity and processivity must be considered when selecting a suitable enzyme for WGA. However, other properties, such as thermostability, capacity to couple replication, and double helix unwinding, or the ability to maintain DNA replication opposite to damaged bases, are also very relevant for some applications.
1. WGA Protocols Based on PCR
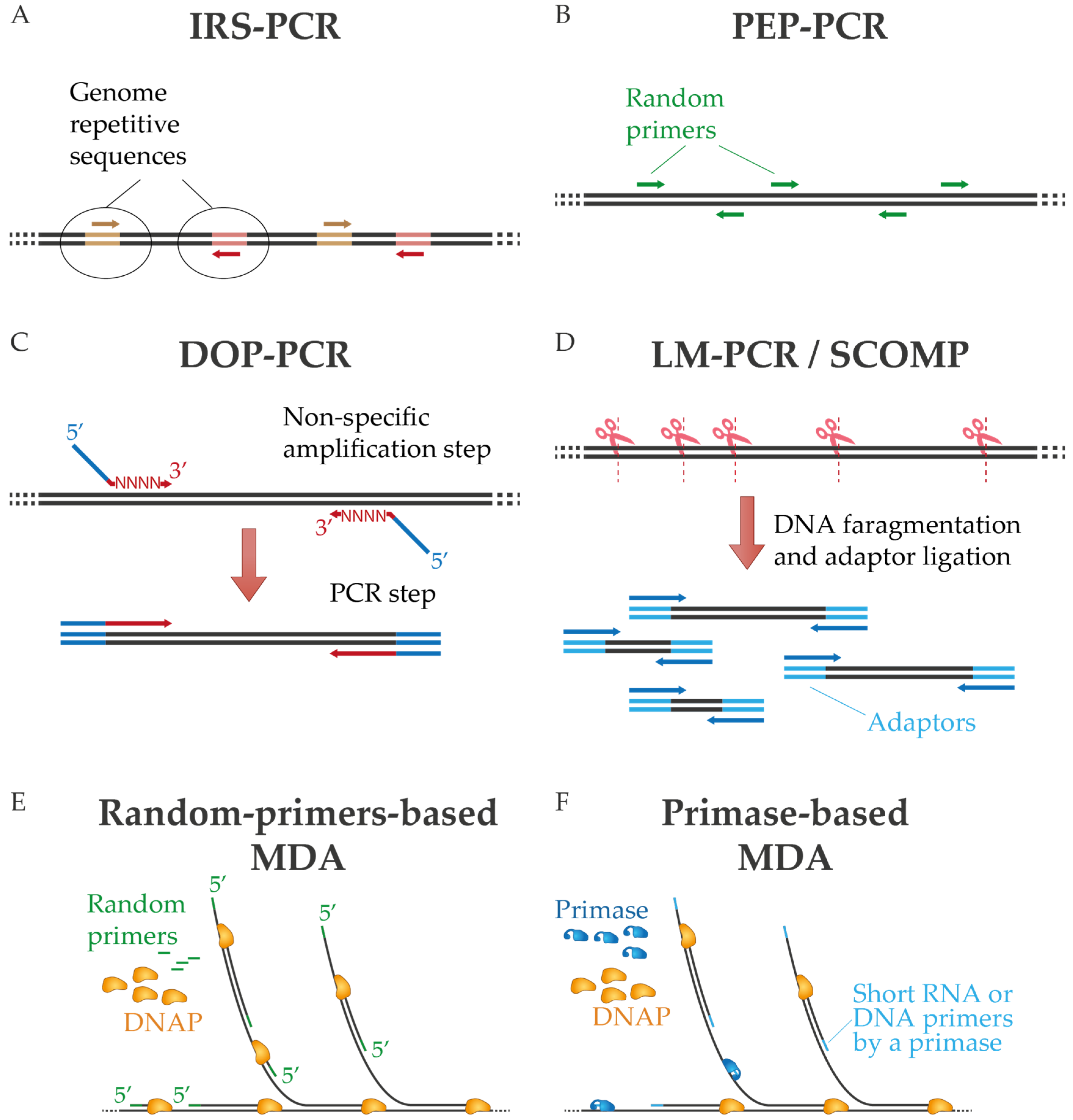
2. WGA Protocols Based on Multiple Displacement Amplification (MDA)
References
- Saiki, R.K.; Scharf, S.; Faloona, F.; Mullis, K.B.; Horn, G.T.; Erlich, H.A.; Arnheim, N. Enzymatic Amplification of β-Globin Genomic Sequences and Restriction Site Analysis for Diagnosis of Sickle Cell Anemia. Science 1985, 230, 1350–1354.
- Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific Enzymatic Amplification of DNA In Vitro: The Polymerase Chain Reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51, 263–273.
- McIver, C.J.; Jacques, C.F.H.; Chow, S.S.W.; Munro, S.C.; Scott, G.M.; Roberts, J.A.; Craig, M.E.; Rawlinson, W.D. Development of Multiplex PCRs for Detection of Common Viral Pathogens and Agents of Congenital Infections. J. Clin. Microbiol. 2005, 43, 5102–5110.
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA-J. Am. Med. Assoc. 2020, 323, 1843–1844.
- Śpibida, M.; Krawczyk, B.; Olszewski, M.; Kur, J. Modified DNA Polymerases for PCR Troubleshooting. J. Appl. Genet. 2017, 58, 133–142.
- Himmelbauer, H.; Schalkwyk, L.C.; Lehrach, H. Interspersed Repetitive Sequence (IRS)-PCR for Typing of Whole Genome Radiation Hybrid Panels. Nucleic Acids Res. 2000, 28, e7.
- Nelson, D.L. Interspersed Repetitive Sequence Polymerase Chain Reaction (IRS PCR) for Generation of Human DNA Fragments from Complex Sources. Methods 1991, 2, 60–74.
- Reyes, G.R.; Kim, J.P. Sequence-Independent, Single-Primer Amplification (SISPA) of Complex DNA Populations. Mol. Cell. Probes 1991, 5, 473–481.
- Froussard, P. RPCR: A Powerful Tool for Random Amplification of Whole RNA Sequences. Genome Res. 1993, 2, 185–190.
- Zou, N.; Ditty, S.; Li, B.; Lo, S.C. Random Priming PCR Strategy to Amplify and Clone Trace Amounts of DNA. BioTechniques 2003, 35, 758–765.
- Brinkmann, A.; Uddin, S.; Krause, E.; Surtees, R.; Dinçer, E.; Kar, S.; Hacıoğlu, S.; Özkul, A.; Ergünay, K.; Nitsche, A. Utility of a Sequence-Independent, Single-Primer-Amplification (SISPA) and Nanopore Sequencing Approach for Detection and Characterization of Tick-Borne Viral Pathogens. Viruses 2021, 13, 203.
- Fouts, D.E. Amplification for Whole Genome Sequencing of Bacteriophages from Single Isolated Plaques Using SISPA. Bacteriophages 2018, 3, 165–178.
- Blagodatskikh, K.A.; Kramarov, V.M.; Barsova, E.V.; Garkovenko, A.V.; Shcherbo, D.S.; Shelenkov, A.A.; Ustinova, V.V.; Tokarenko, M.R.; Baker, S.C.; Kramarova, T.V.; et al. Improved DOP-PCR (IDOP-PCR): A Robust and Simple WGA Method for Efficient Amplification of Low Copy Number Genomic DNA. PLoS ONE 2017, 12, e0184507.
- Telenius, H.; Carter, N.P.; Bebb, C.E.; Nordenskjöld, M.; Ponder, B.A.J.; Tunnacliffe, A. Degenerate Oligonucleotide-Primed PCR: General Amplification of Target DNA by a Single Degenerate Primer. Genomics 1992, 13, 718–725.
- Arneson, N.; Hughes, S.; Houlston, R.; Done, S. Whole-Genome Amplification by Improved Primer Extension Preamplification PCR (I-PEP-PCR). Cold Spring Harb. Protoc. 2008, 3, pdb.prot4921.
- Moghaddaszadeh-Ahrabi, S.; Farajnia, S.; Rahimi-Mianji, G.; Nejati-Javaremi, A. A Short and Simple Improved-Primer Extension Preamplification (I-PEP) Procedure for Whole Genome Amplification (WGA) of Bovine Cells. Anim. Biotechnol. 2012, 23, 24–42.
- Zhang, L.; Cui, X.; Schmitt, K.; Hubert, R.; Navidi, W.; Arnheim, N. Whole Genome Amplification from a Single Cell: Implications for Genetic Analysis. Proc. Natl. Acad. Sci. USA 1992, 89, 5847–5851.
- Tagoh, H.; Cockerill, P.N.; Bonifer, C. In Vivo Genomic Footprinting Using LM–PCR Methods. In Nuclear Reprogramming: Methods and Protocols; Pells, S., Ed.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 2006; pp. 285–314. ISBN 978-1-59745-005-8.
- Arneson, N.; Hughes, S.; Houlston, R.; Done, S. Whole-Genome Amplification by Single-Cell Comparative Genomic Hybridization PCR (SCOMP). Cold Spring Harb. Protoc. 2008, 3, pdb.prot4923.
- Stoecklein, N.H.; Erbersdobler, A.; Schmidt-Kittler, O.; Diebold, J.; Schardt, J.A.; Izbicki, J.R.; Klein, C.A. SCOMP Is Superior to Degenerated Oligonucleotide Primed-Polymerase Chain Reaction for Global Amplification of Minute Amounts of DNA from Microdissected Archival Tissue Samples. Am. J. Pathol. 2002, 161, 43–51.
- Jäger, R. New Perspectives for Whole Genome Amplification in Forensic STR Analysis. Int. J. Mol. Sci. 2022, 23, 7090.
- Volozonoka, L.; Miskova, A.; Gailite, L. Whole Genome Amplification in Preimplantation Genetic Testing in the Era of Massively Parallel Sequencing. Int. J. Mol. Sci. 2022, 23, 4819.
- Bodulev, O.L.; Sakharov, I.Y. Isothermal Nucleic Acid Amplification Techniques and Their Use in Bioanalysis. Biochem. Mosc. 2020, 85, 147–166.
- Huang, L.; Ma, F.; Chapman, A.; Lu, S.; Xie, X.S. Single-Cell Whole-Genome Amplification and Sequencing: Methodology and Applications. Annu. Rev. Genom. Hum. Genet. 2015, 16, 79–102.
- Dean, F.B.; Nelson, J.R.; Giesler, T.L.; Lasken, R.S. Rapid Amplification of Plasmid and Phage DNA Using Phi29 DNA Polymerase and Multiply-Primed Rolling Circle Amplification. Genome Res. 2001, 11, 1095–1099.
- Salas, M.; Blanco, L.; Lázaro, J.M.; De Vega, M. The Bacteriophage Φ29 DNA Polymerase. IUBMB Life 2008, 60, 82–85.
- Blanco, L.; Bernad, A.; Lázaro, J.M.; Martín, G.; Garmendia, C.; Salas, M. Highly Efficient DNA Synthesis by the Phage Φ29 DNA Polymerase: Symmetrical Mode of DNA Replication. J. Biol. Chem. 1989, 264, 8935–8940.
- Asadi, R.; Mollasalehi, H. The Mechanism and Improvements to the Isothermal Amplification of Nucleic Acids, at a Glance. Anal. Biochem. 2021, 631, 114260.
- Nelson, J.R. Random-Primed, Phi29 DNA Polymerase-Based Whole Genome Amplification. Curr. Protoc. Mol. Biol. 2014, 105, 15.13.
- Lage, J.M.; Leamon, J.H.; Pejovic, T.; Hamann, S.; Lacey, M.; Dillon, D.; Segraves, R.; Vossbrinck, B.; González, A.; Pinkel, D.; et al. Whole Genome Analysis of Genetic Alterations in Small DNA Samples Using Hyperbranched Strand Displacement Amplification and Array–CGH. Genome Res. 2003, 13, 294.
- Sabina, J.; Leamon, J.H. Bias in Whole Genome Amplification: Causes and Considerations. Methods Mol. Biol. 2015, 1347, 15–41.
- Li, Y.; Kim, H.-J.; Zheng, C.; Chow, W.H.A.; Lim, J.; Keenan, B.; Pan, X.; Lemieux, B.; Kong, H. Primase-Based Whole Genome Amplification. Nucleic Acids Res. 2008, 36, e79.
- Schaerli, Y.; Stein, V.; Spiering, M.M.; Benkovic, S.J.; Abell, C.; Hollfelder, F. Isothermal DNA Amplification Using the T4 Replisome: Circular Nicking Endonuclease-Dependent Amplification and Primase-Based Whole-Genome Amplification. Nucleic Acids Res. 2010, 38, e201.
- Picher, Á.J.; Budeus, B.; Wafzig, O.; Krüger, C.; García-Gómez, S.; Martínez-Jiménez, M.I.; Díaz-Talavera, A.; Weber, D.; Blanco, L.; Schneider, A. TruePrime Is a Novel Method for Whole-Genome Amplification from Single Cells Based on TthPrimPol. Nat. Commun. 2016, 7, 13296.
- Martínez-Jiménez, M.I.; García-Gómez, S.; Bebenek, K.; Sastre-Moreno, G.; Calvo, P.A.; Díaz-Talavera, A.; Kunkel, T.A.; Blanco, L. Alternative Solutions and New Scenarios for Translesion DNA Synthesis by Human PrimPol. DNA Repair 2015, 29, 127–138.
- Direito, S.O.L.L.; Zaura, E.; Little, M.; Ehrenfreund, P.; Röling, W.F.M.M. Systematic Evaluation of Bias in Microbial Community Profiles Induced by Whole Genome Amplification. Environ. Microbiol. 2014, 16, 643–657.
- Parras-Moltó, M.; Rodríguez-Galet, A.; Suárez-Rodríguez, P.; López-Bueno, A. Evaluation of Bias Induced by Viral Enrichment and Random Amplification Protocols in Metagenomic Surveys of Saliva DNA Viruses. Microbiome 2018, 6, 119.
- Deleye, L.; De Coninck, D.; Dheedene, A.; De Sutter, P.; Menten, B.; Deforce, D.; Van Nieuwerburgh, F. Performance of a TthPrimPol-Based Whole Genome Amplification Kit for Copy Number Alteration Detection Using Massively Parallel Sequencing. Sci. Rep. 2016, 6, 31825.
- Ordóñez, C.D.; Mayoral-Campos, C.; Egas, C.; Redrejo-Rodríguez, M. A Primer-Independent DNA Polymerase-Based Method for Competent Whole-(Meta)Genome Amplification of Intermediate to High GC Sequences. bioRxiv 2023.
- Redrejo-Rodríguez, M.; Ordóñez, C.D.; Berjón-Otero, M.; Moreno-González, J.; Aparicio-Maldonado, C.; Forterre, P.; Salas, M.; Krupovic, M. Primer-Independent DNA Synthesis by a Family B DNA Polymerase from Self-Replicating Mobile Genetic Elements. Cell Rep. 2017, 21, 1574–1587.
- Zong, C.; Lu, S.; Chapman, A.R.; Xie, X.S. Genome-Wide Detection of Single Nucleotide and Copy Number Variations of a Single Human Cell. Science 2012, 338, 1622–1626.





