Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Firas Fafa | -- | 3107 | 2023-06-20 09:00:55 | | | |
| 2 | Camila Xu | Meta information modification | 3107 | 2023-06-20 09:24:12 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Deshotels, L.; Safa, F.M.; Saba, N.S. NOTCH Signaling in Mantle Cell Lymphoma. Encyclopedia. Available online: https://encyclopedia.pub/entry/45834 (accessed on 07 February 2026).
Deshotels L, Safa FM, Saba NS. NOTCH Signaling in Mantle Cell Lymphoma. Encyclopedia. Available at: https://encyclopedia.pub/entry/45834. Accessed February 07, 2026.
Deshotels, Leigh, Firas M. Safa, Nakhle S. Saba. "NOTCH Signaling in Mantle Cell Lymphoma" Encyclopedia, https://encyclopedia.pub/entry/45834 (accessed February 07, 2026).
Deshotels, L., Safa, F.M., & Saba, N.S. (2023, June 20). NOTCH Signaling in Mantle Cell Lymphoma. In Encyclopedia. https://encyclopedia.pub/entry/45834
Deshotels, Leigh, et al. "NOTCH Signaling in Mantle Cell Lymphoma." Encyclopedia. Web. 20 June, 2023.
Copy Citation
The NOTCH signaling pathway is highly conserved, and its dysregulation has been implicated in the pathogenesis of many B cell malignancies including MCL, chronic lymphocytic leukemia (CLL), splenic marginal zone lymphoma (SMZL), follicular lymphoma (FL), diffuse large B cell lymphoma (DLBCL), and multiple myeloma (MM). The NOTCH genes play a critical role in the early and late phases of normal B cell differentiation. In MCL, mutations in the PEST domain stabilize NOTCH proteins, rendering them resistant to degradation, which subsequently results in the upregulation of genes involved in angiogenesis, cell cycle progression, and cell migration and adhesion.
mantle cell lymphoma
MCL
NOTCH1
NOTCH2
MYC
1. Introduction
Mantle cell lymphoma (MCL) is a CD5-positive, CD23-negative B cell malignancy accounting for 6% of all non-Hodgkin lymphomas (NHL) and carries a poor prognosis, with a median survival of only 4–5 years. MCL is characterized by the translocation t(11;14)(q13;q32), which results in the overexpression of the Cyclin-D1 protein [1][2][3]. Cyclin-D1 regulates the cell cycle via promoting the G1 to S transition, and its overexpression results in the uncontrolled cell growth of the malignant clone [2]. While the translocation t(11;14)(q13;q32) is a hallmark for MCL, it is not sufficient for the malignant transformation, and additional genetic insults are required [3]. MCL exhibits considerable genetic diversity when compared to other lymphomas of the B cell lineage; however, the clinical and therapeutic implications of the majority of these mutations are yet to be determined. Although there has not been a single dominant mutation identified in this disease, some of the most commonly mutated genes include ATM, CCND1, UBR5, TP53, BIRC3, NOTCH1, NOTCH2, and TRAF2 [4]. Notably, NOTCH1 and NOTCH2 were found to be mutated in multiple B cell NHLs, including 5–10% of MCL [5][6].
The NOTCH1 and NOTCH2 genes encode transmembrane receptors with distinct functions in cell development [7]. NOTCH1 expression is observed in immature T cells, and its activation directs early hematopoietic progenitors toward the T cell lineage [8]. Conversely, NOTCH2 expression is primarily seen in mature B cells and is essential for the development of splenic marginal zone B cells [9]. Both NOTCH1 and NOTCH2 regulate the expression of several proto-oncogenes, either directly or indirectly, through downstream effectors [10].
The NOTCH signaling pathway is highly conserved, and its dysregulation has been implicated in the pathogenesis of many B cell malignancies including MCL, chronic lymphocytic leukemia (CLL), splenic marginal zone lymphoma (SMZL), follicular lymphoma (FL), diffuse large B cell lymphoma (DLBCL), and multiple myeloma (MM) [11][12][13]. The mutational impact of NOTCH1 is best described in CLL as it represents one of the most frequent single gene alterations found in CLL at diagnosis (5–15% of cases) but increases to 20% in the relapsed setting [14][15]. While the exact impact of NOTCH1 mutation on CLL’s response to chemoimmunotherapy is controversial, it does not seem to impact overall outcomes in the era of targeted therapy. Nonetheless, mutations in NOTCH1 seem to co-occur more frequently with an unmutated IGHV gene [15]. The most common NOTCH1 mutation in CLL results in a premature stop codon affecting the C-terminal portion of the NOTCH1 receptor, leading to an absence of the proline (P), glutamate (E), serine (S), and threonine (T) (PEST) domain, which regulates protein stability and receptor degradation [16]. In addition to mutations in the NOTCH1 gene, CLL can amplify NOTCH1 signaling through the overexpression of the NOTCH1 receptor, taking advantage of the abundance of NOTCH1 ligands in the lymph node (LN) microenvironment [16]. While NOTCH1 pathway alterations play a critical role in CLL biology, mutations in NOTCH2 are prevalent in SMZL, affecting 20% of cases; however, like NOTCH1 mutations in CLL, the majority of NOTCH2 mutations in SMZL affect the C-terminal region PEST domain of the NOTCH2 receptor [7][11]. More recently, dysregulated NOTCH signaling has been implicated in the pathogenesis of MM as well. Several studies have demonstrated that the progression of MM from a monoclonal gammopathy of undetermined significance (MGUS) relies upon the upregulation of the NOTCH1 receptor and its ligand, JAG1 [13][17].
Mutations in both NOTCH1 and NOTCH2 have been identified in MCL and have been associated with adverse features and more aggressive MCL phenotypes such as blastoid and pleomorphic histologies [6][18][19]. In a study conducted by Bea and colleagues, NOTCH1 and NOTCH2 mutations were found in 5.2% and 4.6% of tumors, respectively [6]. A similar study by Kridel and colleagues showed that 11% of the 123 sequenced MCL samples carry a NOTCH1 mutation [18]. While NOTCH1 mutations are more commonly encountered and are similar to those observed in CLL, NOTCH2 mutations have recently been identified in MCL as an alternative and mutually exclusive occurrence [6][7].
2. Description of the NOTCH Signaling Pathway
In humans, four NOTCH genes have been described that encode four separate NOTCH receptors, NOTCH1–4. NOTCH1 and NOTCH2 are the most widely expressed in human tissues, especially at the developmental stage, while NOTCH3 is expressed in vascular smooth muscle and pericytes, and NOTCH4 is expressed in the endothelium [7].
All NOTCH receptors have a similar structure and are composed of a large NOTCH extracellular domain (NECD), a single-pass transmembrane domain (TM), and a small NOTCH intracellular domain (NICD) (Figure 1) [20]. The NECD is composed of multiple epidermal growth factor (EGF)-like repeats, followed by a negative regulatory region (NRR). The EGF-like repeats are responsible for binding to ligands expressed on neighboring cells, leading to NOTCH receptor activation. The number of EGF-like repeats varies between the NOTCH receptors: 36 for both NOTCH1 and NOTCH2, 34 for NOTCH3, and 29 for NOTCH4. The NRR is composed of three Lin12/Notch repeats (LNR) and a heterodimerization domain (HD) and functions as an inhibitor of constitutive NOTCH receptor activation in the absence of ligands (Figure 1 and Figure 2) [7][21]. The TM domain contains cleavage sites recognized by the gamma secretase complex. TM cleavage results in the release of the NICD, allowing for its translocation to the nucleus [22]. The NICD contains a protein-binding RBPJκ-associated molecule (RAM) domain, seven ankyrin repeats (ANK), a NOTCH cytokine response region (NCR), and a transcriptional activation domain (TAD) [23]. This is followed by a C-terminal region containing the PEST domain, which is an important regulator of protein stability (Figure 1) [7].
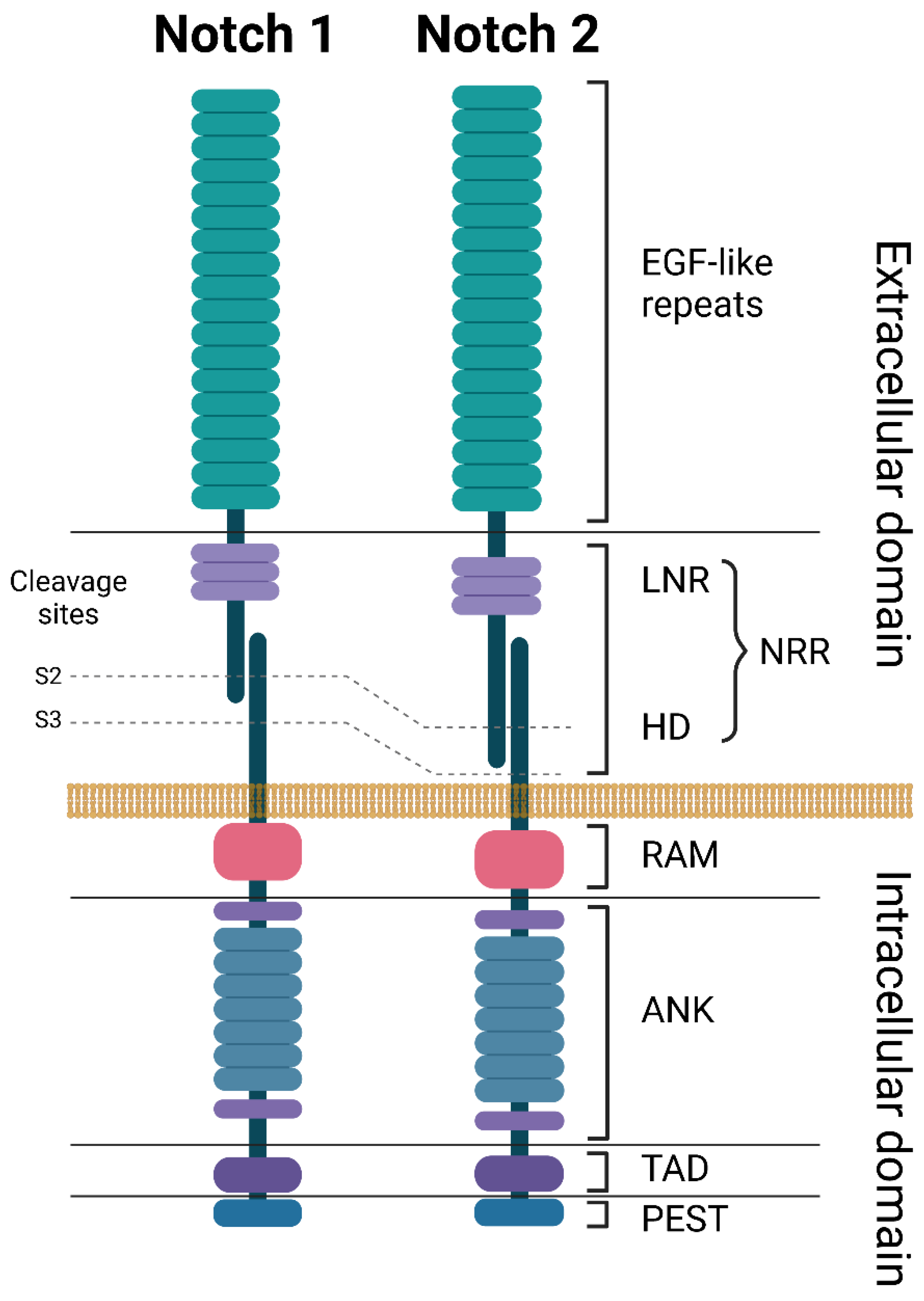
Figure 1. Structures of NOTCH1 and NOTCH2 receptors. ANK: ankyrin repeats, EGF: epidermal growth factor, HD: heterodimerization domain, LNR: Lin-12 Notch repeats, NRR: negative regulatory region, PEST: proline (P), glutamate (E), serine (S), and threonine (T) domain, RAM: RBP-Jkappa-associated module, TAD: transcriptional activation domain. Created with BioRender.com. accessed on 5 June 2023.
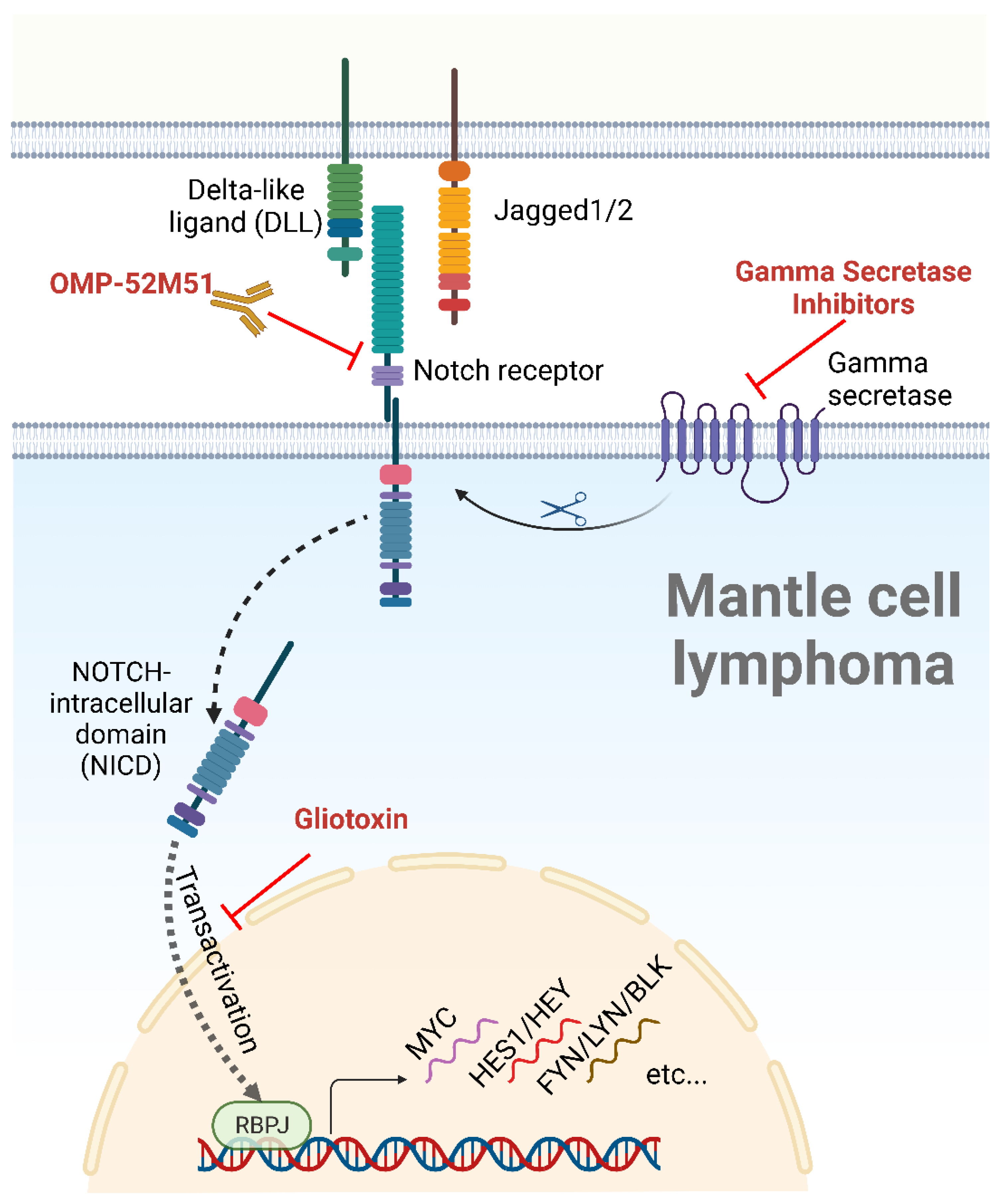
Figure 2. The NOTCH signaling pathway, ligands, and inhibitors. After binding to a cellular ligand, the NOTCH receptor is cleaved by a gamma secretase, allowing the nuclear translocation of the NICD, a process known as transactivation. Following nuclear translocation, the NICD induces the transcription of NOTCH target genes through its interaction with the DNA-binding partner RBPJ. NOTCH inhibitors tested in MCL are depicted in red. Created with BioRender.com, accessed on 5 June 2023.
Cell-to-cell contact is a critical step in the activation of NOTCH pathways as it enables NOTCH receptors to bind to their membrane-bound ligands on neighboring cells (Figure 2) [7][10]. A total of five ligands, grouped into two families, interact with the NOTCH receptors: the Delta-like family (DLL-1, DLL-3, and DLL-4) and the Serrate family of ligands (JAG-1 and JAG-2) (Figure 2) [24]. Interaction with a ligand results in the exposure of the NOTCH receptor’s cleavage site to the protease A-Disintegrin-And-Metalloprotease (ADAM). ADAM-mediated proteolysis leads to the cleavage of the NECD portion of the NOTCH receptor. The TM domain is subsequently cleaved by a gamma secretase complex, releasing the NICD and allowing its translocation to the nucleus (a process called transactivation) [25]. Within the nucleus, The NICD binds to the transcription factor RBPJ and to a mastermind-like (MAML) transcriptional coactivator to form a trimeric complex that activates the transcription of the NOTCH target genes (Figure 2) [26][27].
NOTCH-regulated genes include HES1 and HEY1, transcription repressors that inhibit the expression of genes involved in cell differentiation and apoptosis, DTX1 and NRARP, which provide constitutive NOTCH activation by upregulating a positive feedback loop, and most importantly, MYC, which is a well-known proto-oncogene responsible for the upregulation of cell growth and metabolism [7][28][29].
There are at least two separate mechanisms in which NOTCH activity is terminated. One involves the expression of the HES and HEY family of transcriptional suppressor proteins, which are known targets of NOTCH signaling pathways. Once activated, NOTCH triggers an overexpression of HES and HEY that bind to and inhibit RBPJ, thus creating a negative feedback mechanism, thwarting further NOTCH-induced transcription [29]. Another process involves the degradation of the intracellular portion of the NOTCH receptor via the phosphorylation of the PEST domain [7]. The PEST domain serves as a recognition site for E3 ubiquitin ligases, which covalently attach ubiquitin molecules to lysine residues within the NOTCH receptor [30]. This process marks the intracellular portion of the receptor for recognition and degradation by the proteasome (Figure 2) [30].
3. NOTCH Pathway and Its Role in Normal B Cell Development
The NOTCH1 receptor plays an important role in the early stages of hematopoiesis, specifically in T-cell development. When Pui et al. inoculated progenitor cells transduced with Notch1-espressing retroviral vectors into the bone marrow of mice, they noticed an emergence of a large population of thymic-independent T cells in the bone marrow, in addition to a notable absence of B cell maturation [8]. These and similar findings highly suggest that NOTCH1 activation skews the progenitor cell differentiation toward T cells while suppressing the development of other cell lineages [8][31].
In bone marrow, proper B cell development requires NOTCH1 to be turned off. These antigen-naïve B cells migrate from the bone marrow to secondary lymphoid organs such as the LN and the spleen, where they undergo further maturation through transitional stages (T1 and T2) prior to becoming mature follicular B cells or marginal zone B cells [31].
The interaction between the DLL1 ligand with the NOTCH2 receptor in the marginal zone of the spleen favors marginal zone B cells over follicular B cells [31]. Conversely, Tanigeki and colleagues showed that B cells lacking the downstream transcription factor of NOTCH2, RBPJ, preferentially committed to a follicular cell lineage rather than marginal zone B cell differentiation [32]. These findings highly suggest that activated NOTCH2 in the secondary lymphoid organs is necessary for the B cell transition through the T2 stage. Following that, a continuation of NOTCH2 activation favors marginal zone B cells, while its deactivation favors follicular B cells.
On the other hand, NOTCH1 signaling regulates the differentiation of B cells into antibody-secreting cells. Santos and colleagues stimulated B cells with lipopolysaccharide in the presence of stromal cells transduced with DLL1 [33]. Although the number of B cells recovered was not increased, the antibody production was significantly increased in the DLL1-stimulated B cell population, specifically IgG and IgM [33]. In contrast, the Jagged ligand failed to reproduce these results, suggesting that there is a specificity among the NOTCH ligands with respect to antibody synthesis [34].
In addition to its effects in the spleen, NOTCH signaling plays a critical role in the development of germinal centers (GC) within the LN, where B cells rapidly proliferate and undergo somatic hypermutation, affinity maturation, and isotype switching [34]. NOTCH ligands, specifically DLL1 and JAG1, expressed by follicular dendritic cells in the germinal centers, interact with the NOTCH1 and NOTCH2 receptors expressed on GC B cells [35]. This interaction increases the survival of the GC B cells by regulating the expression of several anti-apoptotic proteins, including Bcl-2 and Mcl-1 [35].
Overall, NOTCH signaling is a critical regulator of B cell development and function, beginning early at the B cell progenitor level and continuing to the end stages of B cell differentiation and antibody production.
4. Cellular Effects of NOTCH Mutations and Their Roles in MCL
Mutations in NOTCH1 and NOTCH2 appear to define a more aggressive MCL subset, such as a frequent association with pleomorphic and blastoid variants and a shorter survival [6][18]. A study by Kridel et al. showed that mutated NOTCH1 represents an independent survival prognostic factor, resulting in a median overall survival (mOS) of 1.4 years compared to 3.8 years in NOTCH1 wild-type cases [18]. In a second report, none of the eight patients carrying a NOTCH2 mutation in their MCL were alive after three years compared to 62% of those without the mutation [6].
While NOTCH mutations in MCL can occur in any location, the PEST domain appears to be the most affected in both NOTCH1 and NOTCH2, harboring more than 85% of the reported mutations (Table 1) [6][18][36][37]. The most common genetic alteration in NOTCH1 is the frame shift alteration P2514Rfs*3 [6][18][36], while the majority of the reported NOTCH2 mutations occurred in exon 34, with the stop-gain R2400* substitution mutation being the most common (Table 1) [6][19]. In both cases, the mutation leads to a truncated PEST domain of the NOTCH protein, resulting in a resistance to E3 ubiquitin ligase-mediated ubiquitination and degradation and thus in a longer half-life of NICD [24].
Table 1. Common NOTCH1 and NOTCH2 mutations in MCL.
| NOTCH1 | NOTCH2 | |||||
|---|---|---|---|---|---|---|
| Mutation | Confirmed Somatic | % of NOTCH1 Mutations † | Mutation | Confirmed Somatic | % of NOTCH2 Mutations † | |
| EGF like repeats | D259N [36] | No | 4 | R91L [37] | Yes | 6.6 |
| P915R [36] | No | 4 | T197I [37] | No | 6.6 | |
| G1088S [18][36] | No | 4 | G225R [37] | Unknown | 6.6 | |
| ANK | R1608H [18] | No | 4 | |||
| D2108N [36] | Yes | 4 | ||||
| PEST | H2428Pfs*7 [18] | Yes | 4 | Q2285* [6] | Yes | 6.6 |
| G2281fs*72 [6] | Unknown | 4 | K2292fs*20 [6] | Unknown | 6.6 | |
| R2431Pfs*5 [18][36] | No | 4 | H2293fs*2 [6] | Unknown | 6.6 | |
| Q2444* [18] | Yes | 4 | P2359A [37] | No | 6.6 | |
| E2460* [18][36] | Yes | 4 | Q2360* [6] | Yes | 6.6 | |
| Q2487* [18] | Yes | 4 | S2391fs*2 [6] | Unknown | 6.6 | |
| V2504fs*3 [6] | Unknown | 4 | R2400* [6][19][37] | Yes | 40 | |
| H2507Pfs*9 [18] | No | 4 | ||||
| P2514Rfs*4 [6][18][36] | Yes | 56 | ||||
† The frequency of each mutation was calculated by dividing the number of each individual mutation by the total number of reported NOTCH mutations (NOTCH1: 25 mutations in 24 samples; NOTCH2: 14 mutations in 14 samples). One sample carried two NOTCH1 mutations (D259N and P2514Rfs*4) [36], and one sample carried a NOTCH1 (P2514Rfs*4) and a NOTCH2 (K2292fs*20) mutation [6].
5. Inhibition of NOTCH Pathways
The inhibition of NOTCH signaling pathways in cancer has proven to be a difficult task, owing in part to the complex mechanism of action of these receptors. Multiple steps are involved in pathway activation, from binding to the ligands to the cleavage of the NICD by gamma secretases, in addition to the effects of NOTCH mutations that induce a ligand-independent pathway activation. Nonetheless, multiple strategies to target NOTCH signaling pathways in cancer have been developed. These strategies include antibodies blocking either extracellular NOTCH domain or its ligands, inhibitors of gamma secretase, decoy agents that bind to the NOTCH receptor, preventing ligand-dependent activation, and agents that target NICD trafficking from the cell membrane to the nucleus [38][39]. Testing of these strategies is ongoing, mostly at the preclinical stage, in a variety of lymphoid cancers, including MCL, CLL, MZL, and T-ALL [40][41][42][43][44].
5.1. NOTCH-Directed Monoclonal Antibodies
The only NOTCH-directed antibody studied in MCL is the humanized anti-NOTCH1 antibody OMP-52M51 (brontictuzumab), a full-length IgG2 humanized monoclonal antibody directed against the LNR and HD domains of NOTCH1. Since brontictuzumab inhibits signaling downstream to NOTCH1 by blocking ligand-dependent receptor activation, its activity is not affected by the presence of PEST domain mutations [45]. The in vitro treatment of DLL4-stimulated Mino cells with brontictuzumab inhibited NOTCH1 activation and abrogated angiogenesis, DNA damage repair, and cellular migration and adhesion. Although brontictuzumab decreased the level of cleaved NOTCH1 in murine MCL xenografts, there was no effect on tumor cell counts and viability [5].
Following encouraging in vitro data, brontictuzumab was tested in a phase 1 clinical trial in 24 patients with relapsed or refractory hematologic malignancies, including 4 patients with MCL [46]. The antibody was relatively well tolerated, with dose-limiting toxicity occurring in two patients. Diarrhea was the most common adverse event, occurring in about one-fifth of patients, followed by fatigue and anemia. Unfortunately, brontictuzumab showed mediocre clinical effects, with only one partial response in a patient with transformed mycosis fungoides and two stable diseases in two others (MCL and transformed mycosis fungoides). Among the three patients who had activating PEST domain mutations in NOTCH1, only one achieved stable disease as a best response [46].
5.2. Gamma Secretase Inhibitors
Gamma secretase inhibitors (GSI) were initially developed for use in Alzheimer’s disease, given the role of gamma secretase in the cleavage of amyloid precursor protein, a key element in disease progression [47][48]. Following discoveries of gamma secretase’s role in cancer, GSIs were repurposed for testing in both solid and hematological malignancies [42][49]. In the preclinical testing of a small-molecule GSI named Compound E on MCL, researchers showed that Compound E significantly decreased Rec-1 and SP-49 cell viabilities at low micromolar concentrations with a concomitant decrease in NICD levels, reflecting a successful inhibitory effect on NOTCH1 cleavage [50].
Among GSIs, the most extensively studied in the clinical sphere to date is RO4929097, a small-molecule inhibitor of gamma secretase. RO4929097 has been tested in phase I and II trials on various solid tumors, including melanoma, glioma, lung, breast, and pancreatic malignancies, and resulted in only a modest clinical benefit [23][51][52][53][54][55]. As an example, when tested in a phase II clinical trial in patients with previously treated metastatic pancreatic cancer, RO4929097 as a single agent failed to induce any objective responses, with only three out of twelve patients achieving a stable disease as the best clinical response, resulting in a median progression-free survival of 1.5 months [56]. A similar phase II of RO4929097 in ovarian cancer resulted only in stable disease as the best clinical response in one-third of the forty-five treated patients and a median progression-free survival of 1.3 months [57]. In both studies, RO4929097 was well tolerated, with fatigue and nausea being the most commonly reported adverse events, while anemia and diarrhea were the most common grade 3 and 4 toxicities, occurring in less than 5% of patients [56][57]. There are currently no clinical trials evaluating GSI in B cell lymphoma.
5.3. Inhibition of NOTCH Transactivation
Gliotoxin is an Aspergillum-derived secondary metabolite that was found to be a potent inhibitor of NOTCH2 transactivation. Hubmann and colleagues showed that gliotoxin exerts an anti-leukemic effect on CLL by inducing apoptosis at the nanomolar range in patient-derived samples, irrespective of the IGHV mutational status or cytogenetic anomalies [44]. The effect of gliotoxin on NOTCH2 signaling resulted in decreased CD23 expression, indicating a suppressive effect on the CD23-encoding gene FCER2, a known target of NOTCH2 [44].
The uniform expression of CD23 in CLL contrasts with its absence in MCL, a pattern used to differentiate between these two CD5-expressing B cell malignancies. Nonetheless, a subset of MCL expresses CD23 with ill-defined causes and consequences. In keeping with the control of NOTCH2 over CD23 expression, the NOTCH2 R2400* mutant MCL cell line Arbo showed a strong CD23 expression which was downregulated by gliotoxin in a dose-dependent manner. Additionally, Arbo cells were highly sensitive to gliotoxin, with an IC50 of 100 nM, while the NOTCH2 wild-type MCL cell lines Jeko-1 and UPN-1 were resistant [19]. To date, there are no clinical trials evaluating gliotoxin in lymphoid malignancies.
Although not tested in MCL, additional ways of inhibiting NOTCH pathways in the preclinical setting include targeting NOTCH degradation and the ICN1 complex. These strategies hold the promise of being more selective to the target, which could translate into improved efficacy and safety profiles [58][59][60][61].
References
- Maddocks, K. Update on mantle cell lymphoma. Blood 2018, 132, 1647–1656.
- Silkenstedt, E.; Dreyling, M. Mantle cell lymphoma-Advances in molecular biology, prognostication and treatment approaches. Hematol. Oncol. 2021, 39 (Suppl. S1), 31–38.
- Saba, N.; Wiestner, A. Do mantle cell lymphomas have an ‘Achilles heel’? Curr. Opin. Hematol. 2014, 21, 350–357.
- Zhang, J.; Jima, D.; Moffitt, A.; Liu, Q.; Czader, M.; Hsi, E.D.; Fedoriw, Y.; Dunphy, C.H.; Richards, K.L.; Gill, J.I.; et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood 2014, 123, 2988–2996.
- Silkenstedt, E.; Arenas, F.; Colom-Sanmartí, B.; Xargay-Torrent, S.; Higashi, M.; Giró, A.; Rodriguez, V.; Fuentes, P.; Aulitzky, W.E.; Van Der Kuip, H.; et al. Notch1 signaling in NOTCH1-mutated mantle cell lymphoma depends on Delta-Like ligand 4 and is a potential target for specific antibody therapy. J. Exp. Clin. Cancer Res. 2019, 38, 1–15.
- Bea, S.; Mas, R.M.V.; Navarro, A.; Salaverria, I.; Martín-Garcia, D.; Jares, P.; Giné, E.; Pinyol, M.; Royo, C.; Nadeu, F.; et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 18250–18255.
- Arruga, F.; Vaisitti, T.; Deaglio, S. The NOTCH Pathway and Its Mutations in Mature B Cell Malignancies. Front. Oncol. 2018, 8, 550.
- Pui, J.C.; Allman, D.; Xu, L.; DeRocco, S.; Karnell, F.G.; Bakkour, S.; Lee, J.Y.; Kadesch, T.; Hardy, R.R.; Aster, J.C.; et al. Notch1 Expression in Early Lymphopoiesis Influences B versus T Lineage Determination. Immunity 1999, 11, 299–308.
- Saito, T.; Chiba, S.; Ichikawa, M.; Kunisato, A.; Asai, T.; Shimizu, K.; Yamaguchi, T.; Yamamoto, G.; Seo, S.; Kumano, K.; et al. Notch2 Is Preferentially Expressed in Mature B Cells and Indispensable for Marginal Zone B Lineage Development. Immunity 2003, 18, 675–685.
- Lobry, C.; Oh, P.; Mansour, M.; Look, A.T.; Aifantis, I. Notch signaling: Switching an oncogene to a tumor suppressor. Blood 2014, 123, 2451–2459.
- Rossi, D.; Trifonov, V.; Fangazio, M.; Bruscaggin, A.; Rasi, S.; Spina, V.; Monti, S.; Vaisitti, T.; Arruga, F.; Famà, R.; et al. The coding genome of splenic marginal zone lymphoma: Activation of NOTCH2 and other pathways regulating marginal zone development. J. Exp. Med. 2012, 209, 1537–1551.
- Tohda, S.; Sato, T.; Kogoshi, H.; Fu, L.; Sakano, S.; Nara, N. Establishment of a novel B-cell lymphoma cell line with suppressed growth by gamma-secretase inhibitors. Leuk. Res. 2006, 30, 1385–1390.
- Nefedova, Y.; Cheng, P.; Alsina, M.; Dalton, W.S.; Gabrilovich, D.I. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood 2004, 103, 3503–3510.
- Puente, X.S.; Pinyol, M.; Quesada, V.; Conde, L.; Ordóñez, G.R.; Villamor, N.; Escaramis, G.; Jares, P.; Beà, S.; González-Díaz, M.; et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011, 475, 101–105.
- Fabbri, G.; Rasi, S.; Rossi, D.; Trifonov, V.; Khiabanian, H.; Ma, J.; Grunn, A.; Fangazio, M.; Capello, D.; Monti, S.; et al. Analysis of the chronic lymphocytic leukemia coding genome: Role of NOTCH1 mutational activation. J. Exp. Med. 2011, 208, 1389–1401.
- Onaindia, A.; Gómez, S.; Piris-Villaespesa, M.; Martínez-Laperche, C.; Cereceda, L.; Montes-Moreno, S.; Batlle, A.; De Villambrosia, S.G.; Pollán, M.; Martín-Acosta, P.; et al. Chronic lymphocytic leukemia cells in lymph nodes show frequent NOTCH1 activation. Haematologica 2015, 100, e200–e203.
- Leong, K.G.; Karsan, A. Recent insights into the role of Notch signaling in tumorigenesis. Blood 2006, 107, 2223–2233.
- Kridel, R.; Meissner, B.; Rogic, S.; Boyle, M.; Telenius, A.; Woolcock, B.; Gunawardana, J.; Jenkins, C.; Cochrane, C.; Ben-Neriah, S.; et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood 2012, 119, 1963–1971.
- Safa, F.; Rasmussen, T.; Lobelle-Rich, P.; Collier, S.; Milligan, N.; Schmeig, J.; Schmid, J.; Wiewiorowski, C.; Totaro, D.; Brown, T.C.; et al. Establishment and characterization of a new mantle cell lymphoma cell line with a NOTCH2 mutation, Arbo. eJHaem 2022, 3, 1326–1329.
- Stanley, P.; Okajima, T. Roles of Glycosylation in Notch Signaling. Curr. Top Dev. Biol. 2010, 92, 131–164.
- Cordle, J.; RedfieldZ, C.; Stacey, M.; van der Merwe, P.A.; Willis, A.C.; Champion, B.R.; Hambleton, S.; Handford, P.A. Localization of the Delta-like-1-binding Site in Human Notch-1 and Its Modulation by Calcium Affinity. J. Biol. Chem. 2008, 283, 11785–11793.
- Mumm, J.S.; Schroeter, E.H.; Saxena, M.T.; Griesemer, A.; Tian, X.; Pan, D.J.; Ray, W.J.; Kopan, R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 2000, 5, 197–206.
- Means-Powell, J.A.; Mayer, I.A.; Ismail-Khan, R.; Del Valle, L.; Tonetti, D.; Abramson, V.G.; Sanders, M.S.; Lush, R.M.; Sorrentino, C.; Majumder, S.; et al. A Phase Ib Dose Escalation Trial of RO4929097 (a gamma-secretase inhibitor) in Combination with Exemestane in Patients with ER + Metastatic Breast Cancer (MBC). Clin. Breast Cancer 2022, 22, 103–114.
- Andersson, E.R.; Sandberg, R.; Lendahl, U. Notch signaling: Simplicity in design, versatility in function. Development 2011, 138, 3593–3612.
- Hubmann, R.; Düchler, M.; Schnabl, S.; Hilgarth, M.; Demirtas, D.; Mitteregger, D.; Hölbl, A.; Vanura, K.; Le, T.; Look, T.; et al. NOTCH2 links protein kinase C delta to the expression of CD23 in chronic lymphocytic leukaemia (CLL) cells. Br. J. Haematol. 2010, 148, 868–878.
- Kurooka, H.; Kuroda, K.; Honjo, T. Roles of the ankyrin repeats and C-terminal region of the mouse Notch1 intracellular region. Nucleic Acids Res. 1998, 26, 5448–5455.
- Giaimo, B.D.; Gagliani, E.K.; Kovall, R.A.; Borggrefe, T. Transcription Factor RBPJ as a Molecular Switch in Regulating the Notch Response. Notch Signal. Embryol. Cancer Notch Signal. Cancer 2020, 1287, 9–30.
- Matsuno, K.; Eastman, D.; Mitsiades, T.; Quinn, A.M.; Carcanciu, M.L.; Ordentlich, P.; Kadesch, T.; Artavanis-Tsakonas, S. Human deltex is a conserved regulator of Notch signalling. Nat. Genet. 1998, 19, 74–78.
- Fischer, A.; Gessler, M. Delta Notch and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 2007, 35, 4583–4596.
- Gupta-Rossi, N.; Le Bail, O.; Gonen, H.; Brou, C.; Logeat, F.; Six, E.; Ciechanover, A.; Israël, A. Functional Interaction between SEL-10, an F-box Protein, and the Nuclear Form of Activated Notch1 Receptor. J. Biol. Chem. 2001, 276, 34371–34378.
- Radtke, F.; Macdonald, H.R.; Tacchini-Cottier, F. Regulation of innate and adaptive immunity by Notch. Nat. Rev. Immunol. 2013, 13, 427–437.
- Tanigaki, K.; Han, H.; Yamamoto, N.; Tashiro, K.; Ikegawa, M.; Kuroda, K.; Suzuki, A.; Nakano, T.; Honjo, T. Notch–RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 2002, 3, 443–450.
- Santos, M.A.; Sarmento, L.M.; Rebelo, M.; Doce, A.A.; Maillard, I.; Dumortier, A.; Neves, H.; Radtke, F.; Pear, W.S.; Parreira, L.; et al. Notch1 engagement by Delta-like-1 promotes differentiation of B lymphocytes to antibody-secreting cells. Proc. Natl. Acad. Sci. USA 2007, 104, 15454–15459.
- Garis, M.; Garrett-Sinha, L.A. Notch Signaling in B Cell Immune Responses. Front. Immunol. 2021, 11, 609324.
- Yoon, S.-O.; Zhang, X.; Berner, P.; Blom, B.; Choi, Y.S. Notch Ligands Expressed by Follicular Dendritic Cells Protect Germinal Center B Cells from Apoptosis. J. Immunol. 2009, 183, 352–358.
- Meissner, B.; Kridel, R.; Lim, R.S.; Rogic, S.; Tse, K.; Scott, D.W.; Moore, R.; Mungall, A.J.; Marra, M.A.; Connors, J.M.; et al. The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood 2013, 121, 3161–3164.
- Saba, N.S.; Liu, D.; Herman, S.E.; Underbayev, C.; Tian, X.; Behrend, D.; Weniger, M.A.; Skarzynski, M.; Gyamfi, J.; Fontan, L.; et al. Pathogenic role of B-cell receptor signaling and canonical NF-kappaB activation in mantle cell lymphoma. Blood 2016, 128, 82–92.
- Roti, G.; Sorrentino, C.; Cuneo, A. Therapeutic targeting of notch signaling pathway in hematological malignancies. Mediterr. J. Hematol. Infect. Dis. 2019, 11, e2019037.
- Espinoza, I.; Miele, L. Notch inhibitors for cancer treatment. Pharmacol. Ther. 2013, 139, 95–110.
- Roti, G.; Qi, J.; Kitara, S.; Sanchez-Martin, M.; Saur Conway, A.; Varca, A.C.; Su, A.; Wu, L.; Kung, A.L.; Ferrando, A.A.; et al. Leukemia-specific delivery of mutant NOTCH1 targeted therapy. J. Exp. Med. 2018, 215, 197–216.
- Real, P.J.; Tosello, V.; Palomero, T.; Castillo, M.; Hernando, E.; de Stanchina, E.; Sulis, M.L.; Barnes, K.; Sawai, C.; Homminga, I.; et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat. Med. 2009, 15, 50–58.
- De Keersmaecker, K.; Lahortiga, I.; Mentens, N.; Folens, C.; Van Neste, L.; Bekaert, S.; Vandenberghe, P.; Odero, M.D.; Marynen, P.; Cools, J. In vitro validation of gamma-secretase inhibitors alone or in combination with other anti-cancer drugs for the treatment of T-cell acute lymphoblastic leukemia. Haematologica 2008, 93, 533–542.
- de Vera Mudry, M.C.; Regenass-Lechner, F.; Ozmen, L.; Altmann, B.; Festag, M.; Singer, T.; Muller, L.; Jacobsen, H.; Flohr, A. Morphologic and functional effects of gamma secretase inhibition on splenic marginal zone B cells. Int. J. Alzheimers Dis. 2012, 2012, 289412.
- Hubmann, R.; Hilgarth, M.; Schnabl, S.; Ponath, E.; Reiter, M.; Demirtas, D.; Sieghart, W.; Valent, P.; Zielinski, C.; Jäger, U.; et al. Gliotoxin is a potent NOTCH2 transactivation inhibitor and efficiently induces apoptosis in chronic lymphocytic leukaemia (CLL) cells. Br. J. Haematol. 2012, 160, 618–629.
- Agnusdei, V.; Minuzzo, S.A.; Frasson, C.; Grassi, A.; Axelrod, F.; Satyal, S.; Gurney, A.; Hoey, T.; Seganfreddo, E.; Basso, G.; et al. Therapeutic antibody targeting of Notch1 in T-acute lymphoblastic leukemia xenografts. Leukemia 2013, 28, 278–288.
- Casulo, C.; Ruan, J.; Dang, N.H.; Gore, L.; Diefenbach, C.; Beaven, A.W.; Castro, J.E.; Porcu, P.; Faoro, L.; Dupont, J.; et al. Safety and Preliminary Efficacy Results of a Phase I First-in-Human Study of the Novel Notch-1 Targeting Antibody Brontictuzumab (OMP-52M51) Administered Intravenously to Patients with Hematologic Malignancies. Blood 2016, 128, 5108.
- Doody, R.S.; Raman, R.; Farlow, M.; Iwatsubo, T.; Vellas, B.; Joffe, S.; Kieburtz, K.; He, F.; Sun, X.; Thomas, R.G.; et al. A phase 3 trial of semagacestat for treatment of Alzheimer’s disease. N. Engl. J. Med. 2013, 369, 341–350.
- Coric, V.; van Dyck, C.H.; Salloway, S.; Andreasen, N.; Brody, M.; Richter, R.W.; Soininen, H.; Thein, S.; Shiovitz, T.; Pilcher, G.; et al. Safety and tolerability of the gamma-secretase inhibitor avagacestat in a phase 2 study of mild to moderate Alzheimer disease. Arch. Neurol. 2012, 69, 1430–1440.
- McCaw, T.R.; Inga, E.; Chen, H.; Jaskula-Sztul, R.; Dudeja, V.; Bibb, J.A.; Ren, B.; Rose, J.B. Gamma Secretase Inhibitors in Cancer: A Current Perspective on Clinical Performance. Oncologist 2020, 26, e608–e621.
- Ryan, R.J.; Petrovic, J.; Rausch, D.M.; Zhou, Y.; Lareau, C.A.; Kluk, M.J.; Christie, A.L.; Lee, W.Y.; Tarjan, D.R.; Guo, B.; et al. A B Cell Regulome Links Notch to Downstream Oncogenic Pathways in Small B Cell Lymphomas. Cell Rep. 2017, 21, 784–797.
- Gold, K.A.; Byers, L.A.; Fan, Y.H.; Fujimoto, J.; Tse, W.H.; Lee, J.J.; Gupta, S.; Wistuba, I.I.; Stewart, D.J.; Gibbons, D.L. A phase I/II trial combining erlotinib with gamma secretase inhibitor RO4929097 in advanced non-small cell lung cancer (NSCLC). J. Clin. Oncol. 2013, 31, 8104.
- Richter, S.; Bedard, P.L.; Chen, E.X.; Clarke, B.A.; Tran, B.; Hotte, S.J.; Stathis, A.; Hirte, H.W.; Razak, A.R.; Reedijk, M.; et al. A phase I study of the oral gamma secretase inhibitor R04929097 in combination with gemcitabine in patients with advanced solid tumors (PHL-078/CTEP 8575). Investig. New Drugs 2014, 32, 243–249.
- Tolcher, A.W.; Messersmith, W.A.; Mikulski, S.M.; Papadopoulos, K.P.; Kwak, E.L.; Gibbon, D.G.; Patnaik, A.; Falchook, G.S.; Dasari, A.; Shapiro, G.I.; et al. Phase I study of RO4929097, a gamma secretase inhibitor of Notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2012, 30, 2348–2353.
- Lee, S.M.; Moon, J.; Redman, B.G.; Chidiac, T.; Flaherty, L.E.; Zha, Y.; Othus, M.; Ribas, A.; Sondak, V.K.; Gajewski, T.F.; et al. Phase 2 study of RO4929097, a gamma-secretase inhibitor, in metastatic melanoma: SWOG 0933. Cancer 2014, 121, 432–440.
- Xu, R.; Shimizu, F.; Hovinga, K.; Beal, K.; Karimi, S.; Droms, L.; Peck, K.K.; Gutin, P.; Iorgulescu, J.B.; Kaley, T.; et al. Molecular and Clinical Effects of Notch Inhibition in Glioma Patients: A Phase 0/I Trial. Clin. Cancer Res. 2016, 22, 4786–4796.
- De Jesus-Acosta, A.; Laheru, D.; Maitra, A.; Arcaroli, J.; Rudek, M.A.; Dasari, A.; Blatchford, P.J.; Quackenbush, K.; Messersmith, W. A phase II study of the gamma secretase inhibitor RO4929097 in patients with previously treated metastatic pancreatic adenocarcinoma. Investig. New Drugs 2014, 32, 739–745.
- Diaz-Padilla, I.; Wilson, M.K.; Clarke, B.; Hirte, H.W.; Welch, S.A.; Mackay, H.J.; Biagi, J.J.; Reedijk, M.; Weberpals, J.I.; Fleming, G.F.; et al. A phase II study of single-agent RO4929097, a gamma-secretase inhibitor of Notch signaling, in patients with recurrent platinum-resistant epithelial ovarian cancer: A study of the Princess Margaret, Chicago and California phase II consortia. Gynecol. Oncol. 2015, 137, 216–222.
- Weber, D.; Lehal, R.; Frismantas, V.; Bourquin, J.-P.; Bauer, M.; Murone, M.; Radtke, F. Pharmacological activity of CB-103: An oral pan-NOTCH inhibitor with a novel mode of action. Ann. Oncol. 2017, 28, v137.
- Spriano, F.; Tarantelli, C.; Arribas, A.; Gaudio, E.; Cascione, L.; Aresu, L.; Zucca, E.; Rossi, D.; Stathis, A.; Murone, M.; et al. Abstract B061: Targeting lymphomas with the novel first-in-class pan-NOTCH transcription inhibitor CB-103. Mol. Cancer Ther. 2018, 17 (Suppl. S1), B061.
- Koyama, D.; Kikuchi, J.; Hiraoka, N.; Wada, T.; Kurosawa, H.; Chiba, S.; Furukawa, Y. Proteasome inhibitors exert cytotoxicity and increase chemosensitivity via transcriptional repression of Notch1 in T-cell acute lymphoblastic leukemia. Leukemia 2013, 28, 1216–1226.
- Duechler, M.; Shehata, M.; Schwarzmeier, J.D.; Hoelbl, A.; Hilgarth, M.; Hubmann, R. Induction of apoptosis by proteasome inhibitors in B-CLL cells is associated with downregulation of CD23 and inactivation of Notch2. Leukemia 2005, 19, 260–267.
More
Information
Subjects:
Hematology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
971
Revisions:
2 times
(View History)
Update Date:
20 Jun 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




