Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Firas Fafa and Version 2 by Camila Xu.
The NOTCH signaling pathway is highly conserved, and its dysregulation has been implicated in the pathogenesis of many B cell malignancies including MCL, chronic lymphocytic leukemia (CLL), splenic marginal zone lymphoma (SMZL), follicular lymphoma (FL), diffuse large B cell lymphoma (DLBCL), and multiple myeloma (MM). The NOTCH genes play a critical role in the early and late phases of normal B cell differentiation. In MCL, mutations in the PEST domain stabilize NOTCH proteins, rendering them resistant to degradation, which subsequently results in the upregulation of genes involved in angiogenesis, cell cycle progression, and cell migration and adhesion.
- mantle cell lymphoma
- MCL
- NOTCH1
- NOTCH2
- MYC
1. Introduction
Mantle cell lymphoma (MCL) is a CD5-positive, CD23-negative B cell malignancy accounting for 6% of all non-Hodgkin lymphomas (NHL) and carries a poor prognosis, with a median survival of only 4–5 years. MCL is characterized by the translocation t(11;14)(q13;q32), which results in the overexpression of the Cyclin-D1 protein [1][2][3][1,2,3]. Cyclin-D1 regulates the cell cycle via promoting the G1 to S transition, and its overexpression results in the uncontrolled cell growth of the malignant clone [2]. While the translocation t(11;14)(q13;q32) is a hallmark for MCL, it is not sufficient for the malignant transformation, and additional genetic insults are required [3]. MCL exhibits considerable genetic diversity when compared to other lymphomas of the B cell lineage; however, the clinical and therapeutic implications of the majority of these mutations are yet to be determined. Although there has not been a single dominant mutation identified in this disease, some of the most commonly mutated genes include ATM, CCND1, UBR5, TP53, BIRC3, NOTCH1, NOTCH2, and TRAF2 [4]. Notably, NOTCH1 and NOTCH2 were found to be mutated in multiple B cell NHLs, including 5–10% of MCL [5][6][5,6].
The NOTCH1 and NOTCH2 genes encode transmembrane receptors with distinct functions in cell development [7]. NOTCH1 expression is observed in immature T cells, and its activation directs early hematopoietic progenitors toward the T cell lineage [8]. Conversely, NOTCH2 expression is primarily seen in mature B cells and is essential for the development of splenic marginal zone B cells [9]. Both NOTCH1 and NOTCH2 regulate the expression of several proto-oncogenes, either directly or indirectly, through downstream effectors [10].
The NOTCH signaling pathway is highly conserved, and its dysregulation has been implicated in the pathogenesis of many B cell malignancies including MCL, chronic lymphocytic leukemia (CLL), splenic marginal zone lymphoma (SMZL), follicular lymphoma (FL), diffuse large B cell lymphoma (DLBCL), and multiple myeloma (MM) [11][12][13][11,12,13]. The mutational impact of NOTCH1 is best described in CLL as it represents one of the most frequent single gene alterations found in CLL at diagnosis (5–15% of cases) but increases to 20% in the relapsed setting [14][15][14,15]. While the exact impact of NOTCH1 mutation on CLL’s response to chemoimmunotherapy is controversial, it does not seem to impact overall outcomes in the era of targeted therapy. Nonetheless, mutations in NOTCH1 seem to co-occur more frequently with an unmutated IGHV gene [15]. The most common NOTCH1 mutation in CLL results in a premature stop codon affecting the C-terminal portion of the NOTCH1 receptor, leading to an absence of the proline (P), glutamate (E), serine (S), and threonine (T) (PEST) domain, which regulates protein stability and receptor degradation [16]. In addition to mutations in the NOTCH1 gene, CLL can amplify NOTCH1 signaling through the overexpression of the NOTCH1 receptor, taking advantage of the abundance of NOTCH1 ligands in the lymph node (LN) microenvironment [16]. While NOTCH1 pathway alterations play a critical role in CLL biology, mutations in NOTCH2 are prevalent in SMZL, affecting 20% of cases; however, like NOTCH1 mutations in CLL, the majority of NOTCH2 mutations in SMZL affect the C-terminal region PEST domain of the NOTCH2 receptor [7][11][7,11]. More recently, dysregulated NOTCH signaling has been implicated in the pathogenesis of MM as well. Several studies have demonstrated that the progression of MM from a monoclonal gammopathy of undetermined significance (MGUS) relies upon the upregulation of the NOTCH1 receptor and its ligand, JAG1 [13][17][13,17].
Mutations in both NOTCH1 and NOTCH2 have been identified in MCL and have been associated with adverse features and more aggressive MCL phenotypes such as blastoid and pleomorphic histologies [6][18][19][6,18,19]. In a study conducted by Bea and colleagues, NOTCH1 and NOTCH2 mutations were found in 5.2% and 4.6% of tumors, respectively [6]. A similar study by Kridel and colleagues showed that 11% of the 123 sequenced MCL samples carry a NOTCH1 mutation [18]. While NOTCH1 mutations are more commonly encountered and are similar to those observed in CLL, NOTCH2 mutations have recently been identified in MCL as an alternative and mutually exclusive occurrence [6][7][6,7].
2. Description of the NOTCH Signaling Pathway
In humans, four NOTCH genes have been described that encode four separate NOTCH receptors, NOTCH1–4. NOTCH1 and NOTCH2 are the most widely expressed in human tissues, especially at the developmental stage, while NOTCH3 is expressed in vascular smooth muscle and pericytes, and NOTCH4 is expressed in the endothelium [7]. All NOTCH receptors have a similar structure and are composed of a large NOTCH extracellular domain (NECD), a single-pass transmembrane domain (TM), and a small NOTCH intracellular domain (NICD) (Figure 1) [20]. The NECD is composed of multiple epidermal growth factor (EGF)-like repeats, followed by a negative regulatory region (NRR). The EGF-like repeats are responsible for binding to ligands expressed on neighboring cells, leading to NOTCH receptor activation. The number of EGF-like repeats varies between the NOTCH receptors: 36 for both NOTCH1 and NOTCH2, 34 for NOTCH3, and 29 for NOTCH4. The NRR is composed of three Lin12/Notch repeats (LNR) and a heterodimerization domain (HD) and functions as an inhibitor of constitutive NOTCH receptor activation in the absence of ligands (Figure 1 and Figure 2) [7][21][7,21]. The TM domain contains cleavage sites recognized by the gamma secretase complex. TM cleavage results in the release of the NICD, allowing for its translocation to the nucleus [22]. The NICD contains a protein-binding RBPJκ-associated molecule (RAM) domain, seven ankyrin repeats (ANK), a NOTCH cytokine response region (NCR), and a transcriptional activation domain (TAD) [23]. This is followed by a C-terminal region containing the PEST domain, which is an important regulator of protein stability (Figure 1) [7].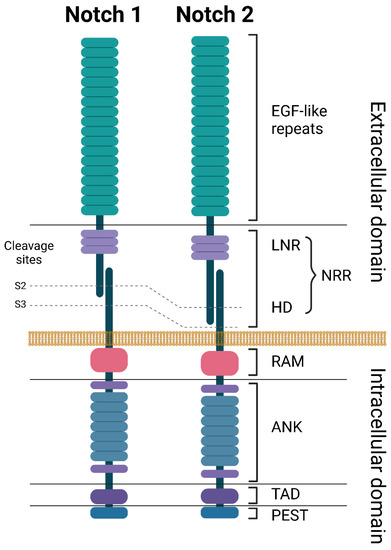
Figure 1. Structures of NOTCH1 and NOTCH2 receptors. ANK: ankyrin repeats, EGF: epidermal growth factor, HD: heterodimerization domain, LNR: Lin-12 Notch repeats, NRR: negative regulatory region, PEST: proline (P), glutamate (E), serine (S), and threonine (T) domain, RAM: RBP-Jkappa-associated module, TAD: transcriptional activation domain. Created with BioRender.com. accessed on 5 June 2023.
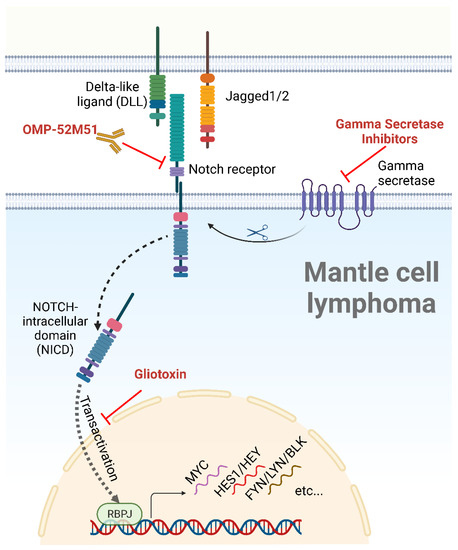
Figure 2. The NOTCH signaling pathway, ligands, and inhibitors. After binding to a cellular ligand, the NOTCH receptor is cleaved by a gamma secretase, allowing the nuclear translocation of the NICD, a process known as transactivation. Following nuclear translocation, the NICD induces the transcription of NOTCH target genes through its interaction with the DNA-binding partner RBPJ. NOTCH inhibitors tested in MCL are depicted in red. Created with BioRender.com, accessed on 5 June 2023.
3. NOTCH Pathway and Its Role in Normal B Cell Development
The NOTCH1 receptor plays an important role in the early stages of hematopoiesis, specifically in T-cell development. When Pui et al. inoculated progenitor cells transduced with Notch1-espressing retroviral vectors into the bone marrow of mice, they noticed an emergence of a large population of thymic-independent T cells in the bone marrow, in addition to a notable absence of B cell maturation [8]. These and similar findings highly suggest that NOTCH1 activation skews the progenitor cell differentiation toward T cells while suppressing the development of other cell lineages [8][31][8,31]. In bone marrow, proper B cell development requires NOTCH1 to be turned off. These antigen-naïve B cells migrate from the bone marrow to secondary lymphoid organs such as the LN and the spleen, where they undergo further maturation through transitional stages (T1 and T2) prior to becoming mature follicular B cells or marginal zone B cells [31]. The interaction between the DLL1 ligand with the NOTCH2 receptor in the marginal zone of the spleen favors marginal zone B cells over follicular B cells [31]. Conversely, Tanigeki and colleagues showed that B cells lacking the downstream transcription factor of NOTCH2, RBPJ, preferentially committed to a follicular cell lineage rather than marginal zone B cell differentiation [32]. These findings highly suggest that activated NOTCH2 in the secondary lymphoid organs is necessary for the B cell transition through the T2 stage. Following that, a continuation of NOTCH2 activation favors marginal zone B cells, while its deactivation favors follicular B cells. On the other hand, NOTCH1 signaling regulates the differentiation of B cells into antibody-secreting cells. Santos and colleagues stimulated B cells with lipopolysaccharide in the presence of stromal cells transduced with DLL1 [33]. Although the number of B cells recovered was not increased, the antibody production was significantly increased in the DLL1-stimulated B cell population, specifically IgG and IgM [33]. In contrast, the Jagged ligand failed to reproduce these results, suggesting that there is a specificity among the NOTCH ligands with respect to antibody synthesis [34]. In addition to its effects in the spleen, NOTCH signaling plays a critical role in the development of germinal centers (GC) within the LN, where B cells rapidly proliferate and undergo somatic hypermutation, affinity maturation, and isotype switching [34]. NOTCH ligands, specifically DLL1 and JAG1, expressed by follicular dendritic cells in the germinal centers, interact with the NOTCH1 and NOTCH2 receptors expressed on GC B cells [35]. This interaction increases the survival of the GC B cells by regulating the expression of several anti-apoptotic proteins, including Bcl-2 and Mcl-1 [35]. Overall, NOTCH signaling is a critical regulator of B cell development and function, beginning early at the B cell progenitor level and continuing to the end stages of B cell differentiation and antibody production.4. Cellular Effects of NOTCH Mutations and Their Roles in MCL
Mutations in NOTCH1 and NOTCH2 appear to define a more aggressive MCL subset, such as a frequent association with pleomorphic and blastoid variants and a shorter survival [6][18][6,18]. A study by Kridel et al. showed that mutated NOTCH1 represents an independent survival prognostic factor, resulting in a median overall survival (mOS) of 1.4 years compared to 3.8 years in NOTCH1 wild-type cases [18]. In a second report, none of the eight patients carrying a NOTCH2 mutation in their MCL were alive after three years compared to 62% of those without the mutation [6]. While NOTCH mutations in MCL can occur in any location, the PEST domain appears to be the most affected in both NOTCH1 and NOTCH2, harboring more than 85% of the reported mutations (Table 1) [6][18][36][37][6,18,36,37]. The most common genetic alteration in NOTCH1 is the frame shift alteration P2514Rfs*3 [6][18][36][6,18,36], while the majority of the reported NOTCH2 mutations occurred in exon 34, with the stop-gain R2400* substitution mutation being the most common (Table 1) [6][19][6,19]. In both cases, the mutation leads to a truncated PEST domain of the NOTCH protein, resulting in a resistance to E3 ubiquitin ligase-mediated ubiquitination and degradation and thus in a longer half-life of NICD [24].Table 1.
Common NOTCH1 and NOTCH2 mutations in MCL.
| NOTCH1 | NOTCH2 | |||||
|---|---|---|---|---|---|---|
| Mutation | Confirmed Somatic | % of NOTCH1 Mutations † | Mutation | Confirmed Somatic | % of NOTCH2 Mutations † | |
| EGF like repeats | D259N [36] | No | 4 | R91L [37] | Yes | 6.6 |
| P915R [36] | No | 4 | T197I [37] | No | 6.6 | |
| G1088S [18][36][18,36] | No | 4 | G225R [37] | Unknown | 6.6 | |
| ANK | R1608H [18] | No | 4 | |||
| D2108N [36] | Yes | 4 | ||||
| PEST | H2428Pfs*7 [18] | Yes | 4 | Q2285* [6] | Yes | 6.6 |
| G2281fs*72 [6] | Unknown | 4 | K2292fs*20 [6] | Unknown | 6.6 | |
| R2431Pfs*5 [18][18[36],36] | No | 4 | H2293fs*2 [6] | Unknown | 6.6 | |
| Q2444* [18] | Yes | 4 | P2359A [37] | No | 6.6 | |
| E2460* [18][36][18,36] | Yes | 4 | Q2360* [6] | Yes | 6.6 | |
| Q2487* [18] | Yes | 4 | S2391fs*2 [6] | Unknown | 6.6 | |
| V2504fs*3 [6] | Unknown | 4 | R2400* [6][19][37][6,19,37] | Yes | 40 | |
| H2507Pfs*9 [18] | No | 4 | ||||
| P2514Rfs*4 [6][18][36][6,18,36] | Yes | 56 | ||||
† The frequency of each mutation was calculated by dividing the number of each individual mutation by the total number of reported NOTCH mutations (NOTCH1: 25 mutations in 24 samples; NOTCH2: 14 mutations in 14 samples). One sample carried two NOTCH1 mutations (D259N and P2514Rfs*4) [36], and one sample carried a NOTCH1 (P2514Rfs*4) and a NOTCH2 (K2292fs*20) mutation [6].
