Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Christos Mikropoulos | -- | 1407 | 2023-06-15 08:21:48 | | | |
| 2 | Conner Chen | Meta information modification | 1407 | 2023-06-19 02:59:19 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Adeleke, S.; Gao, Y.; Okoli, S.; Choi, S.; Ding, H.; Galante, J.R.; Mikropoulos, C. Genomic Alterations in Gynaecological Cancers. Encyclopedia. Available online: https://encyclopedia.pub/entry/45618 (accessed on 12 January 2026).
Adeleke S, Gao Y, Okoli S, Choi S, Ding H, Galante JR, et al. Genomic Alterations in Gynaecological Cancers. Encyclopedia. Available at: https://encyclopedia.pub/entry/45618. Accessed January 12, 2026.
Adeleke, Sola, Yujia Gao, Somto Okoli, Sunyoung Choi, Hao Ding, Joao R. Galante, Christos Mikropoulos. "Genomic Alterations in Gynaecological Cancers" Encyclopedia, https://encyclopedia.pub/entry/45618 (accessed January 12, 2026).
Adeleke, S., Gao, Y., Okoli, S., Choi, S., Ding, H., Galante, J.R., & Mikropoulos, C. (2023, June 15). Genomic Alterations in Gynaecological Cancers. In Encyclopedia. https://encyclopedia.pub/entry/45618
Adeleke, Sola, et al. "Genomic Alterations in Gynaecological Cancers." Encyclopedia. Web. 15 June, 2023.
Copy Citation
Endometrial cancer (EC) and cervical cancer (CC) are common malignancies in women in clinical practice. More uncommon non-ovarian malignancies, such as vulval cancer (VC), are also becoming more prevalent in women of all ages. According to The Cancer Genome Atlas (TCGA) Research Network, ECs can be classified into four groups according to their genetic and molecular information.
endometrial cancer
cervical cancer
vulval cancer
1. Endometrial Cancer
According to The Cancer Genome Atlas (TCGA [1]) Research Network, ECs can be classified into four groups according to their genetic and molecular information (Figure 1):
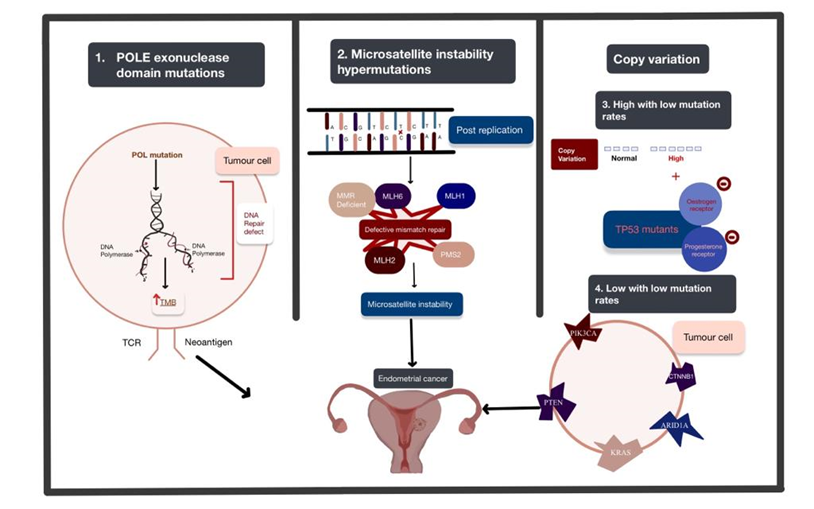
Figure 1. The four genomic alterations expressed in endometrial cancer (EC): POLE exonuclease domain mutations, microsatellite instability hypermutated, copy-number high with low mutation rate, and copy-number low with low mutation rate. The arrows used in this figure depict the order of the progression of these mutations from genetic alteration to the development of EC. Abbreviations: POLE: polymerase Epsilon; POL: polymerase; TMB: tumour mutational burden; TCR: T cell receptor.
Polymerase Epsilon (POLE) ultra-mutated: POLE encodes central catalytic and proofreading subunits of DNA polymerase epsilon involved in leading-strand DNA replication. This subtype is characterised by POLE exonuclease domain mutations (EDMs) mostly in hotspot regions, more than 20% C > A transversions and unusually high mutation rates (232 × 10−6 mutations per Mb). The exonuclease proofreading function locates and replaces erroneous bases in the daughter strand through complementary pairing and ensures a low mutation rate. In endometrial carcinomas, mutations in DNA polymerases inactivate or suppress the proofreading abilities and increase replicative error rates [2]. It is identified in less than 10% [3] of ECs and is associated with aggressive histopathological features but high 5-year recurrence-free survival of around 90% [4].
Microsatellite instability hypermutated (MSI-H): MSI arises from alteration of the MLH1, MSH2, MSH6 and PMS2 genes involved in the post-replicative DNA mismatch repair system. MMR corrects DNA mismatches generated during replication, thereby preventing mutations from becoming permanent [5] in dividing cells, and its defects increase the spontaneous mutation rate [6]. MMR deficiencies can result from inheritance (as in Lynch syndrome), somatic mutations or epigenetic alteration.
The copy-number high with low mutation rate subgroup is characterised by significantly reoccurring amplifications or deletion regions, frequent mutation of TP53, and low oestrogen and progesterone receptor expression. This subgroup shares the same features as basal-like breast cancer and serous ovarian carcinoma.
The copy-number low with low mutation rate subgroup includes the rest of the tumours that do not belong to other groups and are frequently associated with mutations in PTEN, CTNNB1, PIK3CA, ARID1A and KRAS [7]. This subtype appears to share a pathogenesis with colorectal tumours [8].
2. Cervical Cancer
The Cancer Genome Atlas (TCGA) Research Network discovered novel amplifications [9] in immune targets CD274/PD-L1 and PDCD1LG2/PD-L2 and the BCAR4 lncRNA in CCs (Figure 2).
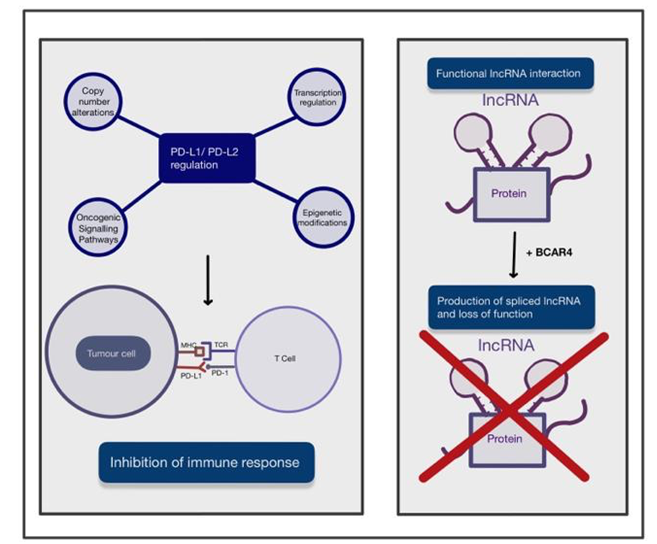
Figure 2. A figure depicting the two main novel amplications in the development of cervical cancer (CC). These include the upregulation of PD-L1/PD-L2 and BCAR4 expression and its role in disrupting the interaction of IncRNA with DNA, RNA and protein complexes. The arrows used in this figure depict the process of these novel amplifications and their effect on immune response or function. Abbreviations: PD-L1: programmed death ligand 1; PD-L2: programmed death ligand 2; MHC: major histocompatibility complex; TCR: tumour cell receptor; BRCA4: breast cancer anti-oestrogen resistance 4.
Programmed cell death ligand 1 (PD-L1) is a receptor mainly expressed on tumour cells and myeloid cells [10]. PD-L1 binds to receptor programmed cell death protein 1 (PD-1) on activated T cells and inhibits the immune response [11] towards tumour cells. PD-L2 is also a ligand of PD-1 and mainly present on antigen-presenting cells [12] and has an overlapping role with PD-L1. The regulation mechanisms of PD-L1/PD-L2 expression are complex and not yet fully understood. PD-L1 expression in tumour cells is regulated by various oncogenic signalling pathways and transcriptional and post-transcriptional factors [13]. Oncogenic factors that induce PD-L1 expression include the accumulation of oncogenic PIK3CA mutations and loss-of-function mutations of the negative regulator in the PI3K/Akt/mTOR pathway. In addition to these, there are also activation of the JAK/STAT pathway and the mitogen-activated protein kinase (MAPK) pathway by genetic mutations and growth factors [13]. Transcription factors, such as HIF-1, STAT3 and NF-κB, can bind to PD-L1 gene promoters, resulting in upregulation of its expression [13]. Finally, post-transcriptional regulation can be disrupted when microRNAs bind to mRNA, resulting in translational repression or alteration [13]. PD-L1 could play a significant role in the future management of CC, as HPV-positivity has been strongly correlated with an increase in PD-1 expression. This has been associated with an improved prognosis and a reduced chance of locoregional recurrence in HPV-positive cancers [14]. Additionally, a correlation has been found between high expression of PD-L1 and increased radiosensitivity, which may contribute to the improved prognostic rates seen in patients with high PD-L1 expression compared to patients with low PD-L1 expression [15]. Copy-number alterations at the 9p24.1 chromosomal region, transcriptional regulation by interferon factors, oncogenic signalling pathways, and epigenetic modifications by enzymes such as histone deacetylases may all contribute to regulation of PD-L1/PD-L2 [13]. In the case of CC, it has been shown that PD-L1 is significantly upregulated in productive HPV infection of the cervix.
Non-coding RNAs (ncRNAs) are RNAs that do not translate into a protein but are responsible for epigenetic regulation [16] at transcriptional and post-transcriptional levels, such as histone modification, DNA methylation and transcriptional silencing. NcRNAs can be categorised by size, and those longer than 200 nucleotides [17] are long ncRNAs (lncRNAs). LncRNAs could drive many important pathological cancer phenotypes [18] through their interactions with other molecules, such as DNA, protein and RNA. For instance, chromatin-bound lncRNAs can regulate gene expression; lncRNA interactions with multiple proteins can promote or impair the assembly of protein complexes; and lncRNAs can recruit proteins involved in mRNA metabolism. The breast cancer anti-oestrogen resistance 4 (BCAR4) gene was previously only studied in the context of breast cancer. BCAR4 produces a spliced lncRNA that is inversely associated with the development of resistance to anti-oestrogens in breast cancer cells and poor disease-free survival for recurrent breast cancer. Therefore, lncRNA BCAR4 was considered to play an important role in the metastasis and tamoxifen-resistance of breast cancer [19]. Over the years, more published evidence has suggested that the prognostic significance of BCAR4 also applies to gastrointestinal malignancy, breast cancer, osteosarcoma and various other cancer types [20]. It has been reported that BCAR4 expression was significantly upregulated in CC tissue and that patients with high BCAR4 expression showed worse survival outcomes. Moreover, overexpression of BCAR4 remarkably promoted the proliferation and motility of CC cells and the epithelial-mesenchymal transition process, while silencing BCAR4 had the reverse effect [21].
3. Vulval Cancer
VCs can develop via two major pathogenetic pathways [22][23], with 30–60% of vulvar tumours being related to HPV infection and the rest having no association with HPV. Both genetic mutations, such as TP53 mutation, and HPV infection can result in similar molecular effects [24], namely, inactivation of p53 and escape from apoptosis. Disruption of the TP53 tumour suppressor gene by point mutations plays a crucial role in VC, with the majority of mutations being single-point mutations in the TP53 core DNA-binding domain [25][26]. The p53 protein is essential for cellular responses under stress, such as DNA damage, hypoxia and oncogene activation. During cellular stresses, p53 binds to DNA as a tetramer by sequence matching and results in gene transcriptional regulation. This process leads to key cellular processes, such as DNA repair, cell-cycle arrest, senescence and apoptosis [27]. Thus, p53 is essential for tumour suppression and can be a potential target for cancer therapy [28].
Other than TP53 mutation as described in Figure 3, it has been reported that PD-L1 expression in squamous cell carcinoma of the vulva could be observed in 32.9% of patients, and its expression in peritumoural immune cells was confirmed in 91.4% of patients [29]. PD-L1 expression upregulation as described in Figure 3, has been suggested to be important for other genomic alterations or modifications described in vulval cancer.
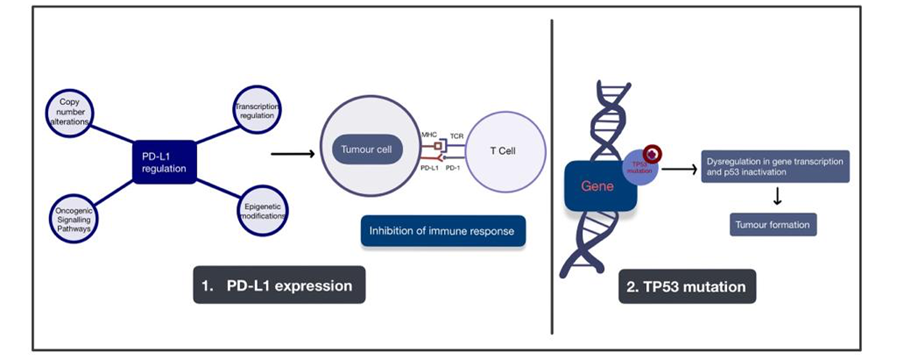
Figure 3. A figure depicting the two major pathogenic pathways involved with the development of vulval cancer. These pathways are the upregulation of PD-L1 expression and mutation of the TP53 gene. Arrows are used to depict the development of tumour formation as a result of these alterations. Abbreviations: PD-L1: programmed death ligand 1; PD-L2: programmed death ligand 2; MHC: major histocompatibility complex; TCR: tumour cell receptor; TP53: tumour protein 53.
Given the extensive knowledge of genomic alterations in EC, CC and VC, it is important that clinicians understand the molecular mechanisms underlying these cancers and use this knowledge to guide approaches to treatment whilst considering approaches to treatment, as well as how these alterations can affect disease progression and treatment efficacy.
References
- Levine, D.A.; Getz, G.; Gabriel, S.B.; Cibulskis, K.; Lander, E.; Sivachenko, A.; Sougnez, C.; Lawrence, M.; Kandoth, C.; Dooling, D.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73.
- Simon, M.; Giot, L.; Faye, G. The 3′ to 5′ exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. Embo J. 1991, 10, 2165–2170.
- Talhouk, A.; McAlpine, J.N. New classification of endometrial cancers: The development and potential applications of genomic-based classification in research and clinical care. Gynecol. Oncol. Res. Pract. 2016, 3, 14.
- León-Castillo, A.; Britton, H.; McConechy, M.K.; McAlpine, J.N.; Nout, R.; Kommoss, S.; Brucker, S.Y.; Carlson, J.W.; Epstein, E.; Rau, T.T.; et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J. Pathol. 2020, 250, 323–335.
- Modrich, P.; Lahue, R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996, 65, 101–133.
- Tiraby, J.G.; Fox, M.S. Marker discrimination in transformation and mutation of pneumococcus. Proc. Natl. Acad. Sci. USA 1973, 70, 3541–3545.
- O’Hara, A.J.; Bell, D.W. The genomics and genetics of endometrial cancer. Adv. Genom. Genet. 2012, 2012, 33–47.
- McConechy, M.K.; Ding, J.; Senz, J.; Yang, W.; Melnyk, N.; Tone, A.A.; Prentice, L.M.; Wiegand, K.C.; McAlpine, J.N.; Shah, S.P.; et al. Ovarian and endometrial endometrioid carcinomas have distinct CTNNB1 and PTEN mutation profiles. Mod. Pathol. 2014, 27, 128–134.
- The Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384.
- Ahmadzadeh, M.; Johnson, L.A.; Heemskerk, B.; Wunderlich, J.R.; Dudley, M.E.; White, D.E.; Rosenberg, S.A. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009, 114, 1537–1544.
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008, 26, 677–704.
- Latchman, Y.; Wood, C.R.; Chernova, T.; Chaudhary, D.; Borde, M.; Chernova, I.; Iwai, Y.; Long, A.J.; Brown, J.A.; Nunes, R.; et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2001, 2, 261–268.
- Zerdes, I.; Matikas, A.; Bergh, J.; Rassidakis, G.Z.; Foukakis, T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: Biology and clinical correlations. Oncogene 2018, 37, 4639–4661.
- Hong, A.M.; Ferguson, P.; Dodds, T.; Jones, D.; Li, M.; Yang, J.; Scolyer, R.A. Significant association of PD-L1 expression with human papillomavirus positivity and its prognostic impact in oropharyngeal cancer. Oral Oncol. 2019, 92, 33–39.
- Lyu, X.; Zhang, M.; Li, G.; Jiang, Y.; Qiao, Q. PD-1 and PD-L1 Expression Predicts Radiosensitivity and Clinical Outcomes in Head and Neck Cancer and is Associated with HPV Infection. J. Cancer 2019, 10, 937–948.
- Costa, F.F. Non-coding RNAs, epigenetics and complexity. Gene 2008, 410, 9–17.
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159.
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016, 29, 452–463.
- Godinho, M.; Meijer, D.; Setyono-Han, B.; Dorssers, L.C.J.; van Agthoven, T. Characterization of BCAR4, a novel oncogene causing endocrine resistance in human breast cancer cells. J. Cell. Physiol. 2011, 226, 1741–1749.
- Zhao, W.; Wang, Z.; Fang, X.; Li, N.; Fang, J. Long noncoding RNA Breast cancer antiestrogen resistance 4 is associated with cancer progression and its significant prognostic value. J. Cell. Physiol. 2019, 234, 12956–12963.
- Zou, R.; Chen, X.; Jin, X.; Li, S.; Ou, R.; Xue, J.; Yan, X.; Chen, L.; Hu, Y.; Zhu, H. Up-regulated BCAR4 contributes to proliferation and migration of cervical cancer cells. Surg. Oncol. 2018, 27, 306–313.
- Smith, J.S.; Backes, D.M.; Hoots, B.E.; Kurman, R.J.; Pimenta, J.M. Human papillomavirus type-distribution in vulvar and vaginal cancers and their associated precursors. Obs. Gynecol. 2009, 113, 917–924.
- van der Avoort, I.A.; Shirango, H.; Hoevenaars, B.M.; Grefte, J.M.; de Hullu, J.A.; de Wilde, P.C.; Bulten, J.; Melchers, W.J.; Massuger, L.F. Vulvar squamous cell carcinoma is a multifactorial disease following two separate and independent pathways. Int. J. Gynecol. Pathol. 2006, 25, 22–29.
- Scheffner, M.; Huibregtse, J.M.; Vierstra, R.D.; Howley, P.M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993, 75, 495–505.
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008.
- Nigro, J.M.; Baker, S.J.; Preisinger, A.C.; Jessup, J.M.; Hostetter, R.; Cleary, K.; Bigner, S.H.; Davidson, N.; Baylin, S.; Devilee, P.; et al. Mutations in the p53 gene occur in diverse human tumour types. Nature 1989, 342, 705–708.
- Riley, T.; Sontag, E.; Chen, P.; Levine, A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell. Biol. 2008, 9, 402–412.
- Vazquez, A.; Bond, E.E.; Levine, A.J.; Bond, G.L. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat. Rev. Drug Discov. 2008, 7, 979–987.
- Thangarajah, F.; Morgenstern, B.; Pahmeyer, C.; Schiffmann, L.M.; Puppe, J.; Mallmann, P.; Hamacher, S.; Buettner, R.; Alidousty, C.; Holz, B.; et al. Clinical impact of PD-L1 and PD-1 expression in squamous cell cancer of the vulva. J. Cancer Res. Clin. Oncol. 2019, 145, 1651–1660.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
721
Revisions:
2 times
(View History)
Update Date:
19 Jun 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




