
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Huikuan Chu | -- | 1984 | 2023-06-08 14:16:25 | | | |
| 2 | Huikuan Chu | -6 word(s) | 1978 | 2023-06-08 14:36:23 | | | | |
| 3 | Lindsay Dong | Meta information modification | 1978 | 2023-06-09 03:37:15 | | |
Video Upload Options
The gut microbiome is made up of bacteria, fungi, viruses and archaea, all of which are closely related with human health. As the main component of enterovirus, the role of bacteriophages (phages) in chronic liver disease has been gradually recognized. Chronic liver diseases, including alcohol-related liver disease and nonalcoholic fatty liver disease, exhibit alterations of the enteric phages. Phages shape the intestinal bacterial colonization and regulate bacterial metabolism. Phages adjoining to intestinal epithelial cells prevents bacteria from invading the intestinal barrier, and mediate intestinal inflammatory response. Phages are also observed increasing intestinal permeability and migrating to peripheral blood and organs, likely contributing to inflammatory injury in chronic liver diseases. By preying on harmful bacteria, phages can improve the gut microbiome of patients with chronic liver disease and thus act as an effective treatment method.
1. Introduction
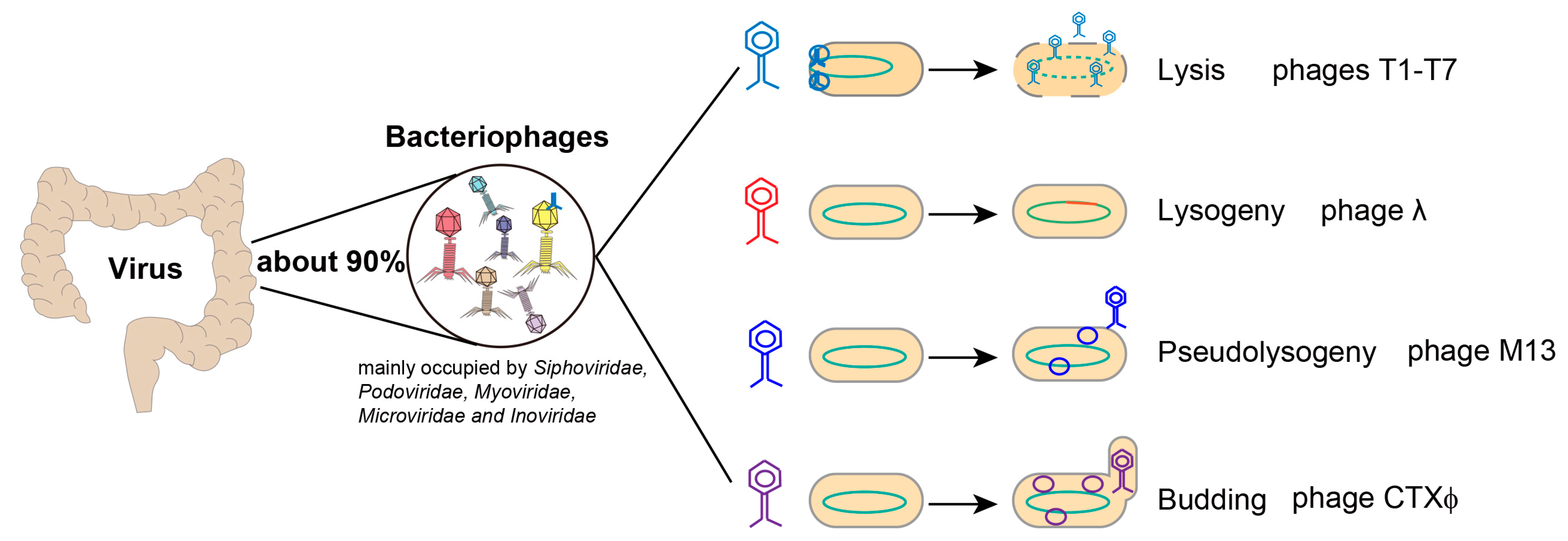
2. Alterations in the Enteric Phages of Chronic Liver Diseases
2.1. Alcoholic Liver Diseases
2.2. Nonalcoholic Fatty Liver Disease
2.3. Liver Cirrhosis
3. Underlying Mechanisms of Phages in Chronic Liver Diseases
3.1. Phages Shape Intestinal Bacterial Colonization
3.2. Phages Regulate Bacterial Metabolism
3.3. Phages and Intestinal Barrier
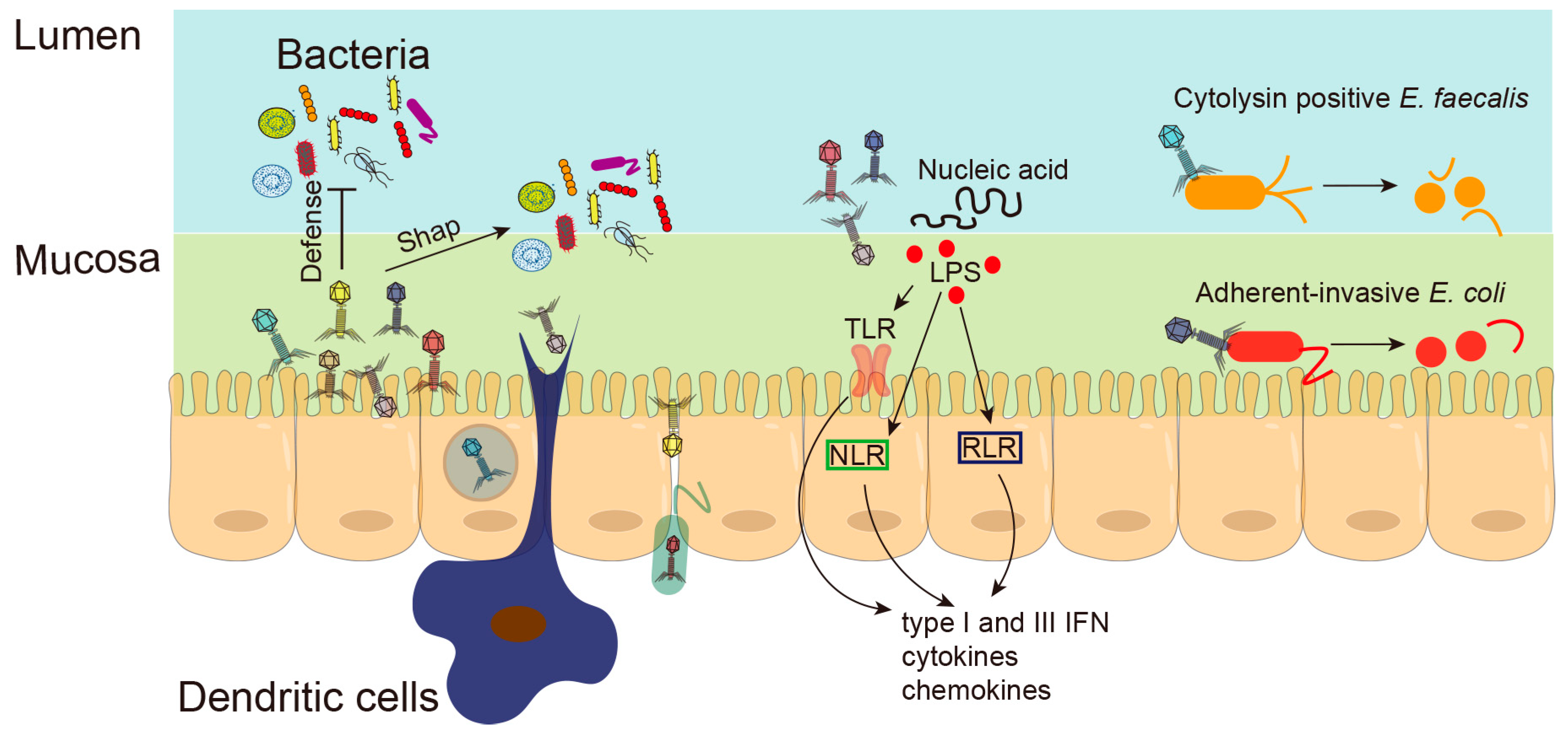
3.4. Phages and Intestinal Inflammation
4. Phage Therapy for Chronic Liver Diseases
References
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138.
- Devoto, A.E.; Santini, J.M.; Olm, M.R.; Anantharaman, K.; Munk, P.; Tung, J.; Archie, E.A.; Turnbaugh, P.J.; Seed, K.D.; Blekhman, R.; et al. Megaphages infect Prevotella and variants are widespread in gut microbiomes. Nat. Microbiol. 2019, 4, 693–700.
- Díaz-Muñoz, S.L.; Koskella, B. Bacteria-phage interactions in natural environments. Adv. Appl. Microbiol. 2014, 89, 135–183.
- Hsu, C.L.; Duan, Y.; Fouts, D.E.; Schnabl, B. Intestinal virome and therapeutic potential of bacteriophages in liver disease. J. Hepatol. 2021, 75, 1465–1475.
- Cao, Z.; Sugimura, N.; Burgermeister, E.; Ebert, M.P.; Zuo, T.; Lan, P. The gut virome: A new microbiome component in health and disease. EBioMedicine 2022, 81, 104113.
- Ma, Y.; You, X.; Mai, G.; Tokuyasu, T.; Liu, C. A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome 2018, 6, 24.
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015, 160, 447–460.
- Hsu, C.L.; Zhang, X.; Jiang, L.; Lang, S.; Hartmann, P.; Pride, D.; Fouts, D.E.; Stärkel, P.; Schnabl, B. Intestinal virome in patients with alcohol use disorder and after abstinence. Hepatol. Commun. 2022, 6, 2058–2069.
- Jiang, L.; Lang, S.; Duan, Y.; Zhang, X.; Gao, B.; Chopyk, J.; Schwanemann, L.K.; Ventura-Cots, M.; Bataller, R.; Bosques-Padilla, F.; et al. Intestinal Virome in Patients With Alcoholic Hepatitis. Hepatology 2020, 72, 2182–2196.
- Fairfield, B.; Schnabl, B. Gut dysbiosis as a driver in alcohol-induced liver injury. JHEP Rep. 2021, 3, 100220.
- Schulfer, A.; Santiago-Rodriguez, T.M.; Ly, M.; Borin, J.M.; Chopyk, J.; Blaser, M.J.; Pride, D.T. Fecal Viral Community Responses to High-Fat Diet in Mice. mSphere 2020, 5, e00833-19.
- Howe, A.; Ringus, D.L.; Williams, R.J.; Choo, Z.-N.; Greenwald, S.M.; Owens, S.M.; Coleman, M.L.; Meyer, F.; Chang, E.B. Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice. ISME J. 2016, 10, 1217–1227.
- Bajaj, J.S.; Sikaroodi, M.; Shamsaddini, A.; Henseler, Z.; Santiago-Rodriguez, T.; Acharya, C.; Fagan, A.; Hylemon, P.B.; Fuchs, M.; Gavis, E.; et al. Interaction of bacterial metagenome and virome in patients with cirrhosis and hepatic encephalopathy. Gut 2021, 70, 1162–1173.
- Fernández, L.; Duarte, A.C.; Rodríguez, A.; García, P. The relationship between the phageome and human health: Are bacteriophages beneficial or harmful microbes? Benef. Microbes 2021, 12, 107–120.
- De Sordi, L.; Lourenço, M.; Debarbieux, L. “I will survive”: A tale of bacteriophage-bacteria coevolution in the gut. Gut Microbes 2019, 10, 92–99.
- Scanlan, P.D. Resistance May Be Futile: Gut Spatial Heterogeneity Supports Bacteria-Phage Co-existence. Cell Host Microbe 2020, 28, 356–358.
- Altuvia, S.; Storz, G.; Papenfort, K. Cross-Regulation between Bacteria and Phages at a Posttranscriptional Level. Microbiol. Spectr. 2018, 6, 4.
- Salmond, G.P.C.; Fineran, P.C. A century of the phage: Past, present and future. Nat. Rev. Microbiol. 2015, 13, 777–786.
- Waldor, M.K.; Mekalanos, J.J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 1996, 272, 1910–1914.
- Ehrbar, K.; Hardt, W.-D. Bacteriophage-encoded type III effectors in Salmonella enterica subspecies 1 serovar Typhimurium. Infect. Genet. Evol. 2005, 5, 1–9.
- Li, Y.; Handley, S.A.; Baldridge, M.T. The dark side of the gut: Virome-host interactions in intestinal homeostasis and disease. J. Exp. Med. 2021, 218, e20201044.
- Li, W.; Wang, A. Genomic islands mediate environmental adaptation and the spread of antibiotic resistance in multiresistant Enterococci—Evidence from genomic sequences. BMC Microbiol. 2021, 21, 55.
- Yu, Z.; Schwarz, C.; Zhu, L.; Chen, L.; Shen, Y.; Yu, P. Hitchhiking Behavior in Bacteriophages Facilitates Phage Infection and Enhances Carrier Bacteria Colonization. Environ. Sci. Technol. 2021, 55, 2462–2472.
- Puxty, R.J.; Millard, A.D. Functional ecology of bacteriophages in the environment. Curr. Opin. Microbiol. 2023, 71, 102245.
- Emerson, J.B.; Roux, S.; Brum, J.R.; Bolduc, B.; Woodcroft, B.J.; Jang, H.B.; Singleton, C.M.; Solden, L.M.; Naas, A.E.; Boyd, J.A.; et al. Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol. 2018, 3, 870–880.
- Shuwen, H.; Kefeng, D. Intestinal phages interact with bacteria and are involved in human diseases. Gut Microbes 2022, 14, 2113717.
- Hsu, B.B.; Gibson, T.E.; Yeliseyev, V.; Liu, Q.; Lyon, L.; Bry, L.; Silver, P.A.; Gerber, G.K. Dynamic Modulation of the Gut Microbiota and Metabolome by Bacteriophages in a Mouse Model. Cell Host Microbe 2019, 25, 803–814.e5.
- Barr, J.J. A bacteriophages journey through the human body. Immunol. Rev. 2017, 279, 106–122.
- Górski, A.; Wazna, E.; Dabrowska, B.-W.; Dabrowska, K.; Switała-Jeleń, K.; Miedzybrodzki, R. Bacteriophage translocation. FEMS Immunol. Med. Microbiol. 2006, 46, 313–319.
- Tetz, G.; Tetz, V. Bacteriophage infections of microbiota can lead to leaky gut in an experimental rodent model. Gut Pathog. 2016, 8, 33.
- Krut, O.; Bekeredjian-Ding, I. Contribution of the Immune Response to Phage Therapy. J. Immunol. 2018, 200, 3037–3044.
- Barr, J.J.; Auro, R.; Furlan, M.; Whiteson, K.L.; Erb, M.L.; Pogliano, J.; Stotland, A.; Wolkowicz, R.; Cutting, A.S.; Doran, K.S.; et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 10771–10776.
- Federici, S.; Nobs, S.P.; Elinav, E. Phages and their potential to modulate the microbiome and immunity. Cell. Mol. Immunol. 2021, 18, 889–904.
- Tetz, G.V.; Ruggles, K.V.; Zhou, H.; Heguy, A.; Tsirigos, A.; Tetz, V. Bacteriophages as potential new mammalian pathogens. Sci. Rep. 2017, 7, 7043.
- Nguyen, S.; Baker, K.; Padman, B.S.; Patwa, R.; Dunstan, R.A.; Weston, T.A.; Schlosser, K.; Bailey, B.; Lithgow, T.; Lazarou, M.; et al. Bacteriophage Transcytosis Provides a Mechanism to Cross Epithelial Cell Layers. mBio 2017, 8, e01874-17.
- Metzger, R.N.; Krug, A.B.; Eisenächer, K. Enteric Virome Sensing-Its Role in Intestinal Homeostasis and Immunity. Viruses 2018, 10, 146.
- Górski, A.; Międzybrodzki, R.; Borysowski, J.; Dąbrowska, K.; Wierzbicki, P.; Ohams, M.; Korczak-Kowalska, G.; Olszowska-Zaremba, N.; Łusiak-Szelachowska, M.; Kłak, M.; et al. Phage as a modulator of immune responses: Practical implications for phage therapy. Adv. Virus Res. 2012, 83, 41–71.
- Gogokhia, L.; Buhrke, K.; Bell, R.; Hoffman, B.; Brown, D.G.; Hanke-Gogokhia, C.; Ajami, N.J.; Wong, M.C.; Ghazaryan, A.; Valentine, J.F.; et al. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host Microbe 2019, 25, 282–299.e8.
- Dąbrowska, K.; Miernikiewicz, P.; Piotrowicz, A.; Hodyra, K.; Owczarek, B.; Lecion, D.; Kaźmierczak, Z.; Letarov, A.; Górski, A. Immunogenicity studies of proteins forming the T4 phage head surface. J. Virol. 2014, 88, 12551–12557.
- Rasmussen, T.S.; Koefoed, A.K.; Jakobsen, R.R.; Deng, L.; Castro-Mejía, J.L.; Brunse, A.; Neve, H.; Vogensen, F.K.; Nielsen, D.S. Bacteriophage-mediated manipulation of the gut microbiome—Promises and presents limitations. FEMS Microbiol. Rev. 2020, 44, 507–521.
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18.
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu. Rev. Med. 2022, 73, 197–211.
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232.
- Kakasis, A.; Panitsa, G. Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Int. J. Antimicrob. Agents 2019, 53, 16–21.
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733.
- Yen, M.; Cairns, L.S.; Camilli, A. A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat. Commun. 2017, 8, 14187.
- Zuo, T.; Wong, S.H.; Lam, K.; Lui, R.; Cheung, K.; Tang, W.; Ching, J.Y.L.; Chan, P.K.S.; Chan, M.C.W.; Wu, J.C.Y.; et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut 2018, 67, 634–643.
- Park, H.; Laffin, M.R.; Jovel, J.; Millan, B.; Hyun, J.E.; Hotte, N.; Kao, D.; Madsen, K.L. The success of fecal microbial transplantation in Clostridium difficile infection correlates with bacteriophage relative abundance in the donor: A retrospective cohort study. Gut Microbes 2019, 10, 676–687.
- Duan, Y.; Young, R.; Schnabl, B. Bacteriophages and their potential for treatment of gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 135–144.
- Sabino, J.; Hirten, R.P.; Colombel, J.-F. Review article: Bacteriophages in gastroenterology-from biology to clinical applications. Aliment. Pharmacol. Ther. 2020, 51, 53–63.
- Galtier, M.; De Sordi, L.; Sivignon, A.; de Vallée, A.; Maura, D.; Neut, C.; Rahmouni, O.; Wannerberger, K.; Darfeuille-Michaud, A.; Desreumaux, P.; et al. Bacteriophages Targeting Adherent Invasive Escherichia coli Strains as a Promising New Treatment for Crohn’s Disease. J. Crohn’s Colitis 2017, 11, 840–847.
- Hesse, S.; Malachowa, N.; Porter, A.R.; Freedman, B.; Kobayashi, S.D.; Gardner, D.J.; Scott, D.P.; Adhya, S.; DeLeo, F.R. Bacteriophage Treatment Rescues Mice Infected with Multidrug-Resistant Klebsiella pneumoniae ST258. mBio 2021, 12, e00034-21.
- Duan, Y.; Llorente, C.; Lang, S.; Brandl, K.; Chu, H.; Jiang, L.; White, R.C.; Clarke, T.H.; Nguyen, K.; Torralba, M.; et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 2019, 575, 505–511.




