Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Emmanuel Nii Adjei Annan | -- | 1969 | 2023-06-06 16:41:17 | | | |
| 2 | Lindsay Dong | Meta information modification | 1969 | 2023-06-08 05:17:03 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Annan, E.N.; Huang, L. Mechanisms of Co-Evolution of Wheat and Rust Pathogens. Encyclopedia. Available online: https://encyclopedia.pub/entry/45250 (accessed on 08 February 2026).
Annan EN, Huang L. Mechanisms of Co-Evolution of Wheat and Rust Pathogens. Encyclopedia. Available at: https://encyclopedia.pub/entry/45250. Accessed February 08, 2026.
Annan, Emmanuel N., Li Huang. "Mechanisms of Co-Evolution of Wheat and Rust Pathogens" Encyclopedia, https://encyclopedia.pub/entry/45250 (accessed February 08, 2026).
Annan, E.N., & Huang, L. (2023, June 06). Mechanisms of Co-Evolution of Wheat and Rust Pathogens. In Encyclopedia. https://encyclopedia.pub/entry/45250
Annan, Emmanuel N. and Li Huang. "Mechanisms of Co-Evolution of Wheat and Rust Pathogens." Encyclopedia. Web. 06 June, 2023.
Copy Citation
Wheat (Triticum spp.) is a cereal crop domesticated >8000 years ago and the second-most-consumed food crop nowadays. Ever since mankind has written records, cereal rust diseases have been a painful awareness in antiquity documented in the Old Testament (about 750 B.C.). The pathogen causing the wheat stem rust disease is among the first identified plant pathogens in the 1700s, suggesting that wheat and rust pathogens have co-existed for thousands of years. With advanced molecular technologies, wheat and rust genomes have been sequenced, and interactions between the host and the rust pathogens have been extensively studied at molecular levels.
wheat
rust
effector
co-evolution
1. History of Wheat-Rust Co-Existence
Wheat refers to the cultivated Triticum spp., including the most common hexaploid bread wheat (T. aestivum L.) and the tetraploid durum wheat (T. turgidum var. durum). These cereal crops are the staple food in most developing countries and are the most traded grains globally. Wheat rust is among the earliest documented plant diseases, dating back to Aristotle’s time (384–322 B.C.) [1]. Epidemics of rust diseases were a reason for an ancient practice of Robigus, the rust god [2], and now the diseases are still one of the major constraints for wheat production worldwide [3]. There are three fungal species from the genus of Puccinia, P. triticina (Pt), P. graminis f. sp. tritici (Pgt), and P. striiformis f. sp. tritici (Pst) causing leaf, stem and stripe rust on wheat, respectively. The Pgt was first described with details by two Italian scientists, Fontana and Tozzetti, independently in 1767 and named by Persoon in 1797 [1]. Anton De Bary found barberry as an alternative host of Pgt in 1865 [1]. The three species of rusts are obligate biotrophic parasites that require living host cells to grow and reproduce [4]. However, the parasites can remain alive as spores in the absence of a living host for a period of time depending on the conditions [5]. Five types of spores are produced by the three rust pathogens, production of urediniospores, and teliospores on the primary grass hosts such as wheat by asexual reproduction. Teliospores germinate to produce basidiospores infect an alternative host to form pycniospores (spermatia) and produce aeciospores by sexual reproduction on the alternative host [6][7].
2. No Interactions during Rust Germination
Rust infections on their primary grass hosts, including wheat, start from aeciospores/urediniospores landing on the leaf surfaces. Under moist conditions, free water is required for spore germination, a process that occurs at night in the natural environment. Expression analysis using a cDNA library from germinated Pst urediniospores revealed germination stage-specific-expressed genes [8]. Over 60% of the genes were involved in primary metabolism (42.6%) and protein synthesis (21.6%). Some of the stage-specific-expressed Pst genes shared significant homology with known virulence factors such as HESP767 of flax rust and PMK1, GAS1, and GAS2 of rice blast fungus [8].
The germination of spores does not require living hosts, and the process can happen on a plastic surface when free water is present [8]. However, germination is inhibited by endogenous self-inhibitors released from the spores if the population density floated on the water is too high [9]. Each rust species has its optimum germination temperature in the range of 11~20 °C. Neither host genotypes (resistant or susceptible) nor host extracts [10] affect spore germination rates, thus suggesting that host defense responses did not happen at the stage of spore germination before penetration [11].
3. Molecular Interactions during Penetration and Haustoria Formation
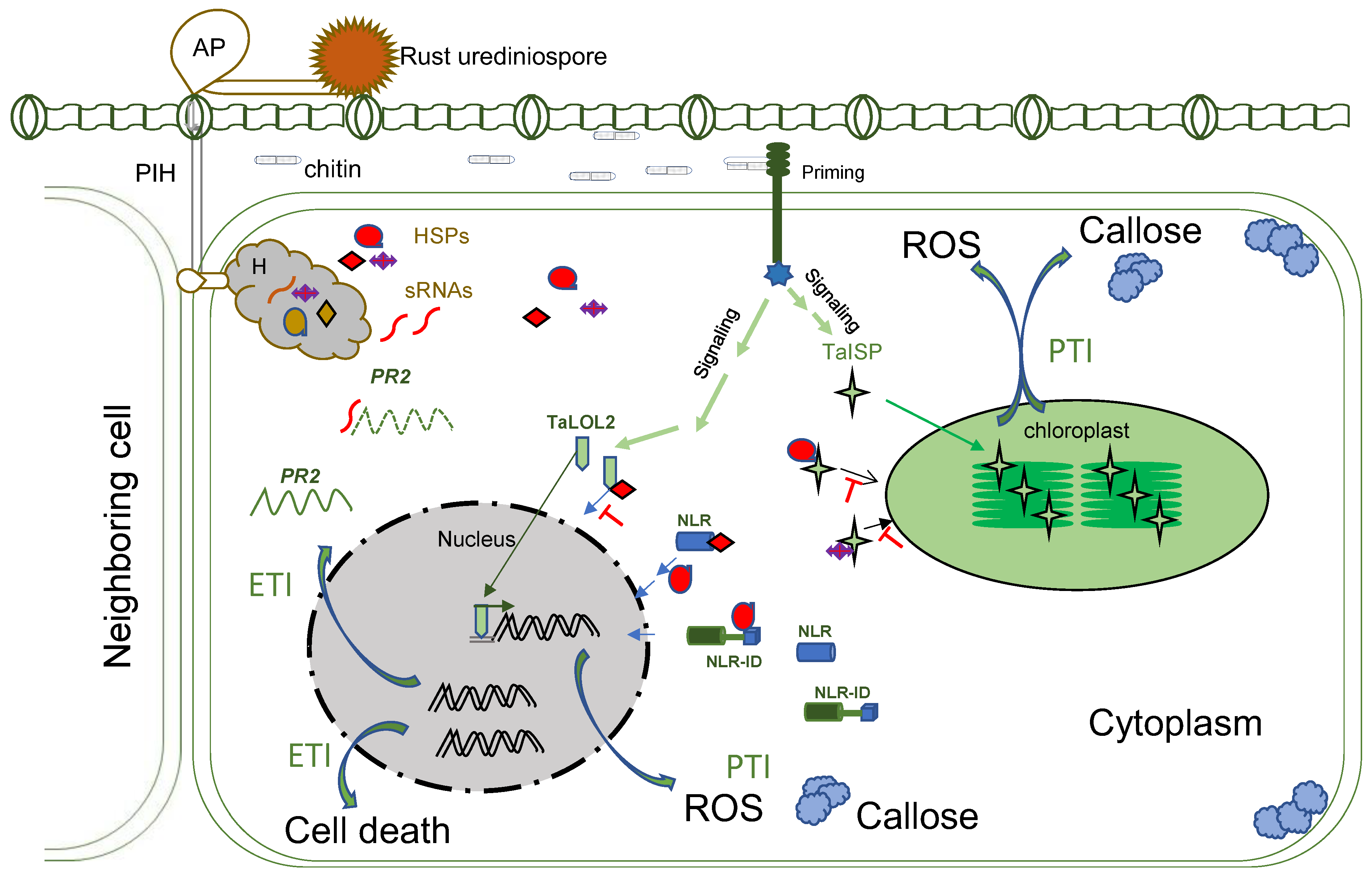
After germination, the germ tubes grow perpendicular to leaf veins until they encounter stomata. The topology of specific host guard cells plays a vital role in stomata identification, known as thigmotropism. Formation of an appressorium is induced over a stoma around 4~16 h post-germination [12]. Then, an appressorium forms a penetration peg to initiate penetration, and a substomatal tube grows between the two guard cells of the host, a mechanism previously determined to be light-dependent [13]. Penetration is shortly followed by substomatal growth, including the formation of primary infection hyphae (PIH) that grow intracellularly until they encounter mesophyll cells. Once a PIH contacts a living mesophyll cell, the tip of the PIH differentiates to a haustorial mother cell (HMC), and a haustorium is induced (Figure 1). The HMC located outside of the mesophyll cell degrades a tiny hole on the cell wall and invaginate the plasma membrane to form an intracellular young haustorium [14] at about 24 h post-infection (hpi), and a mature haustorium as early as 48 hpi [15]. If the host cell in contact with the PIH is dying, the PIH differentiation stops, and hyphae will continue to grow to find another viable cell [15]. The mechanism to govern this process is still unknown.

Figure 1. Molecular interactions with wheat host during rust infection. AP: appressorium; PIH: primary infection hyphae; H: haustorium; HSPs: haustorial secreted proteins; sRNA: small RNA; PR2: pathogenesis-related protein 2; ROS: Reactive Oxygen Species; TaISP: Triticum aestivum iron-sulfur protein; TaLOLs: Triticum aestivum LSD-One-Like 2; NLR: nucleotide-binding and leucine-rich repeat; NLR-ID: NLR–integrated domain; PTI: pathogenic-associated molecular pattern-triggered immunity; ETI: effector-triggered immunity.
During penetration of the host cell, a fungal cell wall component chitin can be an elicitor to trigger the wheat defense response [16], with the signatures of bursts of reactive oxygen species (ROS) such as H2O2 and increased callose deposition around the penetration sites [17][18], known as pathogenic-associated molecular pattern-triggered immunity (PTI). The host PTI is regulated by priming, signal cascades, and movements of transcription factors to the nuclear to activate more genes or movements of nucleus-encoded proteins to chloroplasts to generate ROS and callose. Wheat transcription factors TaLOL2, TaCBF1d, and a copper–zinc superoxide dismutase TaCZSOD2 are known to be positively involved in PTI [19][20][21].
At the early penetration stage, rusts deploy effectors to interfere with different steps of host PTI operated by the nucleus or organelles, such as chloroplasts. For example, the Pst effector PstGSRE1, a glycine–serine-rich protein, disrupted the movement of TaLOL2 to the nucleus [20]. TaLOL2 is a transcription factor promoting ROS. PstGSRE4 targeted the wheat copper–zinc superoxide dismutase TaCZSOD2, an enzyme positively involved in PTI [19]. The PstPEC6 effector interacted with the wheat adenosine kinase to hamper ROS accumulation and callose deposition [22]. Genes encoding PEC6 homologs are conserved among Pst isolates and highly similar among three rust species [23], suggesting a conserved strategy for suppressing PTI among the rusts. Pst effector PsSpg1 interacted with TaPslPK1 to promote its nuclear localization and subsequent phosphorylation of TaCBF1d and degradation. TaCBF1d is a crucial transcription factor in activating PTI [21]. Inactivation of TaPslPK1 rendered wheat with broad-spectrum resistance to rusts [21].
Once the host PTI was suppressed, rust could form haustoria, and haustorial-specific/enriched genes could start to express or upregulate [24] as an effector repertoire. Transcriptomic studies of multiple isolates of each Pst, Pgt, and Pt revealed that the Pst had 1989 differential expressed genes in haustoria; 400 possess a secretion peptide, and >40% of the genes are involved in metabolic processes and translation [25], all six Pt races had 456 haustorial secreted proteins (HSPs) [26], and four different Pgt isolates had 520 HSPs [27].
In addition to the deployment of effectors to attack crucial components of PTI/ effector-triggered immunity (ETI), rust produces small RNAs (sRNA) as important pathogenicity factors to impair host immunity [28]. For example, A 21-nt microRNA of Pst (Pst-milR1) was found to bind the wheat PR2 gene to reduce the transcript abundance of the gene and suppress the host defense during infection. PR2 is a β-1, 3-glucanase with antifungal property [29]. High production of PR2 is the result of active ETI. Silencing the precursor of the Pst-milR1 resulted in wheat resistance to the pathogen [28]. A transcriptomic study on wheat–Pst interaction at 7 dpi discovered differential expression profiles of sRNAs from wheat and Pst [30]. More 35-nt and less 24-nt sRNAs from the Pst infection wheat than the uninfected plant. Pst produced abundant sRNAs almost entirely of 19–23-nt in sizes [30].
Accordingly, wheat has a sophisticated surveillance system to detect rust effectors and activates even stronger defense responses, known as ETI. For example, wheat Sr35 detects Pgt AvrSr35 and mediates resistance to stop the PIH growth before the formation of haustoria [31]. So far, there are 231 designated rust resistance genes [32][33][34][35][36][37], including 83 leaf rust resistance (Lr) genes, 64 stem rust resistance (Sr) genes and 84 stripe rust resistance (Yr) genes. So many rust resistance genes implicate a high variation from their counterpart.
The first cloned wheat rust resistance gene Lr21 [38] shed light on the defense mechanism of wheat ETI against leaf rust. Lr21 encodes a protein with nucleotide-binding and leucine-rich repeat (NLR) domains, mediating resistance with hypersensitive cell death and high pathogenesis-related protein (PR) productions. Up to now, 29 rust resistance genes have been cloned [39][40][41][42]. Twenty-three of the 29 R proteins (79%) belong to the NLR class. After the bread wheat whole genome was sequenced, sequence annotation revealed that the wheat genome contains ~2151 NLR-like genes [43]. These genes are arranged in 547 gene clusters and located at the distal ends of the chromosomes, known as recombination hotspots [43][44].
The NLR-like proteins identified in the wheat genome are classified as the majority to be the classical NLRs containing an N-terminal coiled-coil (CC)/TIR + NB + LRR domains [42] and some NLR fusion proteins, e.g., an NLR fused with integrated domain (ID) homologous with proteins of different functions [42][45][46]. The abundance and locations of the wheat NLR proteins and their structures left footprints on how the new genes were evolved, and what the possible modes of their defense actions are.
Classical NLRs are known to recognize pathogen effectors, also called avirulence (Avr) proteins, directly or indirectly [47]. Studies on the molecular interactions of three pairs of wheat Sr proteins and the matching Avr proteins, Sr35/AvrSr35 [31], Sr50/AvrSr50 [48], and Sr27/AvrSr27 [40], demonstrated that mutations at the DNA or expression level of the matching effector genes were the mechanisms of generating new Pgt virulence isolates. For example, the Sr35-mediated resistance stops the development of PIH before the formation of haustoria [31]. A transposon-mediated insertion in AvrSr35 created Sr35-virulent Pgt isolates with increased expression in a susceptible wheat line [31].
There are 28 different types of IDs that have been identified to be present in either the N- or C-terminus of an NLR in wheat [43]. These IDs include kinase and DNA-binding domains, which function as signal transduction. An ID of a kinase or a DNA-binding in an NLR may help the NLR to initiate defense signaling through phosphorylation of transcription factors by the biochemical activity of kinase [49] or move directly to the nucleus with DNA-binding to the promoters of genes involved in defense responses. However, an ID such as NPR1 fused with an NLR protein is likely to serve as a molecular bait or decoy. NPR1 is a key transcriptional regulator in plant defense [50][51][52][53][54]. Wheat has 6 members of NPR1-like genes located in homoeologous groups 3 and 7. The group 3 NPR1 genes regulate the salicylic acid (SA)-signaling pathway and the crosstalk between SA- and Jasmonic acid (JA)- pathways. The Ta7ANPR1 locus in wheat encodes two types of NLRs, NB-ARC, and NB-ARC + NPR1, through alternative splicing [46]. Together with a CC + NB-ARC gene at a head-to-head orientation, it monitors the integrity of NPR1 proteins. Mutations in the Ta7ANPR1 activated resistance to rusts [46].
4. Molecular Interactions during Colonization and Nutrient Acquisition
After all the battles with wheat, ultimately, rust pathogens want to obtain nutrients from the host to grow and reproduce. Haustoria are believed to be the powerhouse of feeding sites for pathogen colony growth. The biotrophic nature and lifestyle of the rusts imply that the pathogen needs to manipulate the host cellular machinery to redirect the sink sources to haustorial sites. Understanding of the molecular interactions during this step is still limited. A wheat gene Lr34 encoding an ABC transporter confers durable and non-specific resistance against biotrophic fungal pathogens [55]. Lr34res and Lr34sus differ by two nucleotides, and as a result, the LR34sus protein was undetectable in planta due to post-transcriptional regulation [56]. The substrate of the LR34 ABC transporter is phytohormone abscisic acid (ABA), the redistribution of ABA by LR34 has been found to have a major effect on the transcriptional response and physiology of the host, resulting in resistance [56].
References
- Schumann, G.; Leonard, K. Stem rust of wheat (black rust). Plant Health Instr. 2000.
- Zadoks, J.C. Cereal rusts, dogs and stars in antiquity. Cereal Rusts Bull. 1985, 13, 1–10.
- Singh, J.; Chhabra, B.; Raza, A.; Yang, S.H.; Sandhu, K.S. Important wheat diseases in the US and their management in the 21st century. Front. Plant Sci. 2022, 13, 1010191.
- Kolmer, J.A.; Ordonez, M.E.; Groth, J.V. The Rust Fungi. In ELS, 1st ed.; John Wiley & Sons, Ltd., Ed.; Wiley: Hoboken, NJ, USA, 2009; ISBN 978-0-470-01617-6.
- Maddison, A.C.; Manners, J.G. Sunlight and viability of cereal rust uredospores. Trans. Br. Mycol. Soc. 1972, 59, 429–443.
- Bushnell, W.R.; Roelfs, A.P. (Eds.) The Cereal Rusts; Academic Press: Orlando, FL, USA, 1984; ISBN 978-0-12-148401-9.
- Green, G.J.; Johnson, T.; Conners, I.L. Pioneer Leaders in Plant Pathology: J.H. Craigie. Annu. Rev. Phytopathol. 1980, 18, 19–25.
- Zhang, Y.; Qu, Z.; Zheng, W.; Liu, B.; Wang, X.; Xue, X.; Xu, L.; Huang, L.; Han, Q.; Zhao, J.; et al. Stage-specific gene expression during urediniospore germination in Puccinia striiformis f. sp. tritici. BMC Genom. 2008, 9, 203.
- Macko, V.; Staples, R.C.; Allen, P.J.; Renwick, J.A.A. Identification of the germination self-inhibitor from wheat stem rust uredospores. Science 1971, 173, 835–836.
- Anderson, J.A. Studies on the nature of rust resistance in wheat: VI. effect of hydrogen ion concentration, phenolic compounds, and host extracts on the germination of urediniospores of puccinia graminis tritici, form 21. Can. J. Res. 1934, 11, 667–686.
- Jacobs, T. Germination and appressorium formation of wheat leaf rust on susceptible, partially resistant and resistant wheat seedlings and on seedlings of other Gramineae. Neth. J. Plant Pathol. 1989, 95, 65–71.
- Figueroa, M.; Upadhyaya, N.M.; Sperschneider, J.; Park, R.F.; Szabo, L.J.; Steffenson, B.; Ellis, J.G.; Dodds, P.N. Changing the game: Using integrative genomics to probe virulence mechanisms of the stem rust pathogen Puccinia graminis f. sp. tritici. Front. Plant Sci. 2016, 7, 205.
- Yirgou, D.; Caldwell, R.M. Stomatal penetration of wheat seedlings by stem and leaf rust: Effect of light and carbon dioxide. Science 1963, 141, 272–273.
- Solanki, S.; Ameen, G.; Borowicz, P.; Brueggeman, R.S. Shedding Light on penetration of cereal host stomata by wheat stem rust using improved methodology. Sci. Rep. 2019, 9, 7939.
- Talajoor, M.; Wang, X.; Zhang, H.; Zhang, P.; Bhavani, S.; Tabe, L.; Lagudah, E.; Huang, L. Wheat mutant MNR220 delays haustoria formation during leaf rust pathogenesis at the seedling stage. Can. J. Plant Pathol. 2016, 38, 338–347.
- Su, Y.; Chen, Y.; Chen, J.; Zhang, Z.; Guo, J.; Cai, Y.; Zhu, C.; Li, Z.; Zhang, H. Effectors of Puccinia striiformis f. sp. tritici suppressing the pathogenic-associated molecular pattern-triggered immune response were screened by transient expression of wheat protoplasts. Int. J. Mol. Sci. 2021, 22, 4985.
- Cheng, Y.; Wang, X.; Yao, J.; Voegele, R.T.; Zhang, Y.; Wang, W.; Huang, L.; Kang, Z. Characterization of protein kinase PsSRPKL, a novel pathogenicity factor in the wheat stripe rust fungus. Environ. Microbiol. 2015, 17, 2601–2617.
- Cheng, Y.; Wu, K.; Yao, J.; Li, S.; Wang, X.; Huang, L.; Kang, Z. PSTha5a23, a candidate effector from the obligate biotrophic pathogen Puccinia striiformis f. sp. tritici, is involved in plant defense suppression and rust pathogenicity. Environ. Microbiol. 2017, 19, 1717–1729.
- Liu, C.; Wang, Y.; Wang, Y.; Du, Y.; Song, C.; Song, P.; Yang, Q.; He, F.; Bai, X.; Huang, L.; et al. Glycine-serine-rich effector PstGSRE4 in Puccinia striiformis f. sp. tritici inhibits the activity of copper zinc superoxide dismutase to modulate immunity in wheat. PLoS Pathog. 2022, 18, e1010702.
- Qi, T.; Guo, J.; Liu, P.; He, F.; Wan, C.; Islam, M.A.; Tyler, B.M.; Kang, Z.; Guo, J. Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-promoting transcription factor TaLOL2 to defeat ROS-induced defense in wheat. Mol. Plant 2019, 12, 1624–1638.
- Wang, N.; Tang, C.; Fan, X.; He, M.; Gan, P.; Zhang, S.; Hu, Z.; Wang, X.; Yan, T.; Shu, W.; et al. Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 2022, 185, 2961–2974.e19.
- Liu, C.; Pedersen, C.; Schultz-Larsen, T.; Aguilar, G.B.; Madriz-Ordeñana, K.; Hovmøller, M.S.; Thordal-Christensen, H. The stripe rust fungal effector PEC6 suppresses pattern-triggered immunity in a host species-independent manner and interacts with adenosine kinases. New Phytol. 2016.
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 2010, 22, 3130–3141.
- Xu, J.; Linning, R.; Fellers, J.; Dickinson, M.; Zhu, W.; Antonov, I.; Joly, D.L.; Donaldson, M.E.; Eilam, T.; Anikster, Y.; et al. Gene discovery in EST sequences from the wheat leaf rust fungus Puccinia triticina sexual spores, asexual spores and haustoria, compared to other rust and corn smut fungi. BMC Genom. 2011, 12, 161.
- Garnica, D.P.; Upadhyaya, N.M.; Dodds, P.N.; Rathjen, J.P. Strategies for wheat stripe rust pathogenicity identified by transcriptome sequencing. PLoS ONE 2013, 8, e67150.
- Bruce, M.; Neugebauer, K.; Joly, D.; Migeon, P.; Cuomo, C.; Wang, S.; Akhunov, E.; Bakkeren, G.; Kolmer, J.; Fellers, J. Using transcription of six Puccinia triticina races to identify the effective secretome during infection of wheat. Front. Plant Sci. 2014, 4, 520.
- Upadhyaya, N.M.; Garnica, D.P.; Karaoglu, H.; Sperschneider, J.; Nemri, A.; Xu, B.; Mago, R.; Cuomo, C.A.; Rathjen, J.P.; Park, R.F.; et al. Comparative genomics of Australian isolates of the wheat stem rust pathogen Puccinia graminis f. sp. tritici reveals extensive polymorphism in candidate effector genes. Front. Plant Sci. 2014, 5, 759.
- Wang, B.; Sun, Y.; Song, N.; Zhao, M.; Liu, R.; Feng, H.; Wang, X.; Kang, Z. Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol. 2017, 215, 338–350.
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1,3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012, 34, 1983–1990.
- Mueth, N.A.; Hulbert, S.H. Small RNAs target native and cross-kingdom transcripts on both sides of the wheat stripe rust interaction. Genomics 2022, 114, 110526.
- Salcedo, A.; Rutter, W.; Wang, S.; Akhunova, A.; Bolus, S.; Chao, S.; Anderson, N.; De Soto, M.F.; Rouse, M.; Szabo, L.; et al. Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science 2017, 358, 1604–1606.
- McIntosh, R.; Mu, J.; Han, D.; Kang, Z. Wheat stripe rust resistance gene Yr24/Yr26: A retrospective review. Crop J. 2018, 6, 321–329.
- Kumar, S.; Bhardwaj, S.C.; Gangwar, O.P.; Sharma, A.; Qureshi, N.; Kumaran, V.V.; Khan, H.; Prasad, P.; Miah, H.; Singh, G.P.; et al. Lr80: A new and widely effective source of leaf rust resistance of wheat for enhancing diversity of resistance among modern cultivars. Theor. Appl. Genet. 2021, 134, 849–858.
- Kolmer, J.A.; Bajgain, P.; Rouse, M.N.; Li, J.; Zhang, P. Mapping and characterization of the recessive leaf rust resistance gene Lr83 on wheat chromosome arm1DS. Theor. Appl. Genet. 2023, 136, 115.
- Mcintosh, R.; Yamazaki, Y.; Dubcovsky, J.; Rogers, J.; Morris, C.; Xia, X. Catalogue of gene symbols for wheat. Annu. Wheat Newsl. 2022, 68, 72–77.
- Mago, R.; Chen, C.; Xia, X.; Whan, A.; Forrest, K.; Basnet, B.; Perera, G.; Chandramohan, S.; Randhawa, M.; Hayden, M.; et al. Adult plant stem rust resistance in durum wheat glossy huguenot: Mapping, marker development and validation. Appl. Genet. 2022, 135, 1541–1550.
- Klymiuk, V.; Chawla, H.S.; Wiebe, K.; Ens, J.; Fatiukha, A.; Govta, L.; Fahima, T.; Pozniak, C. Discovery of stripe rust resistance with incomplete dominance in wild emmer wheat using bulked segregant analysis sequencing. Commun. Biol. 2022, 5, 1–9.
- Huang, L.; Brooks, S.A.; Li, W.; Fellers, J.P.; Trick, H.N.; Gill, B.S. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 2003, 164, 655–664.
- Luo, J.; Rouse, M.N.; Hua, L.; Li, H.; Li, B.; Li, T.; Zhang, W.; Gao, C.; Wang, Y.; Dubcovsky, J.; et al. Identification and characterization of Sr22b, a new allele of the wheat stem rust resistance gene Sr22 effective against the Ug99 race group. Plant Biotechnol. J. 2022, 20, 554–563.
- Upadhyaya, N.M.; Mago, R.; Panwar, V.; Hewitt, T.; Luo, M.; Chen, J.; Sperschneider, J.; Nguyen-Phuc, H.; Wang, A.; Ortiz, D.; et al. Genomics accelerated isolation of a new stem rust avirulence gene–wheat resistance gene pair. Nat. Plants 2021, 7, 1220–1228.
- Yu, G.; Matny, O.; Champouret, N.; Steuernagel, B.; Moscou, M.J.; Hernández-Pinzón, I.; Green, P.; Hayta, S.; Smedley, M.; Harwood, W.; et al. Aegilops sharonensis genome-assisted identification of stem rust resistance gene Sr62. Nat. Commun. 2022, 13, 1607.
- Zhang, J.; Zhang, P.; Dodds, P.; Lagudah, E. How target-sequence enrichment and sequencing (TEnSeq) pipelines have catalyzed resistance gene cloning in the wheat-rust pathosystem. Front. Plant Sci. 2020, 11, 678.
- Andersen, E.J.; Nepal, M.P.; Purintun, J.M.; Nelson, D.; Mermigka, G.; Sarris, P.F. Wheat disease resistance genes and their diversification through integrated domain fusions. Front. Genet. 2020, 11, 898.
- International Wheat Genome Sequencing Consortium (IWGSC) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191.
- Bailey, P.C.; Schudoma, C.; Jackson, W.; Baggs, E.; Dagdas, G.; Haerty, W.; Moscou, M.; Krasileva, K.V. Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 2018, 19, 23.
- Wang, X.; Zhang, H.; Nyamesorto, B.; Luo, Y.; Mu, X.; Wang, F.; Kang, Z.; Lagudah, E.; Huang, L. A new mode of NPR1 action via an NB-ARC–NPR1 fusion protein negatively regulates the defence response in wheat to stem rust pathogen. New Phytol. 2020, 228, 959–972.
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814.
- Chen, J.; Upadhyaya, N.M.; Ortiz, D.; Sperschneider, J.; Li, F.; Bouton, C.; Breen, S.; Dong, C.; Xu, B.; Zhang, X.; et al. Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science 2017, 358, 1607–1610.
- Sarris, P.F.; Cevik, V.; Dagdas, G.; Jones, J.D.G.; Krasileva, K.V. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 2016, 14, 8.
- Cao, H.; Bowling, S.A.; Gordon, A.S.; Dong, X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 1994, 6, 1583–1592.
- Dong, X. NPR1, all things considered. Curr. Opin. Plant Biol. 2004, 7, 547–552.
- Pajerowska-Mukhtar, K.M.; Emerine, D.K.; Mukhtar, M.S. Tell me more: Roles of NPRs in plant immunity. Trends Plant Sci. 2013, 18, 402–411.
- Rate, D.N.; Greenberg, J.T. The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J. 2001, 27, 203–211.
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.C.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Brown, R.; Kazan, K.; et al. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 2003, 15, 760–770.
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323, 1360–1363.
- Krattinger, S.G.; Kang, J.; Bräunlich, S.; Boni, R.; Chauhan, H.; Selter, L.L.; Robinson, M.D.; Schmid, M.W.; Wiederhold, E.; Hensel, G.; et al. Abscisic acid is a substrate of the ABC transporter encoded by the durable wheat disease resistance gene Lr34. New Phytol. 2019, 223, 853–866.
More
Information
Subjects:
Others
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
908
Revisions:
2 times
(View History)
Update Date:
08 Jun 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




