
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hiroki Kuniyasu | -- | 2242 | 2023-05-24 12:03:38 | | | |
| 2 | Lindsay Dong | Meta information modification | 2242 | 2023-05-25 02:52:31 | | |
Video Upload Options
Claudin-4 (CLDN4) is a key component of tight junctions (TJs) in epithelial cells. CLDN4 is overexpressed in many epithelial malignancies and correlates with cancer progression. Changes in CLDN4 expression have been associated with epigenetic factors (such as hypomethylation of promoter DNA), inflammation associated with infection and cytokines, and growth factor signaling. CLDN4 helps to maintain the tumor microenvironment by forming TJs and acts as a barrier to the entry of anticancer drugs into tumors. Decreased expression of CLDN4 is a potential marker of epithelial-mesenchymal transition (EMT), and decreased epithelial differentiation due to reduced CLDN4 activity is involved in EMT induction. Non-TJ CLDN4 also activates integrin beta 1 and YAP to promote proliferation, EMT, and stemness. These roles in cancer have led to investigations of molecular therapies targeting CLDN4 using anti-CLDN4 extracellular domain antibodies, gene knockdown, clostridium perfringens enterotoxin (CPE), and C-terminus domain of CPE (C-CPE), which have demonstrated the experimental efficacy of this approach.
1. Introduction
2. CLDN4 Expression and Regulation in Cancer
2.1. Epigenetics
2.2. Inflammatory Processes
2.3. Growth Factors
3. The Function of CLDN4 in Cancer
3.1. Carcinogenesis
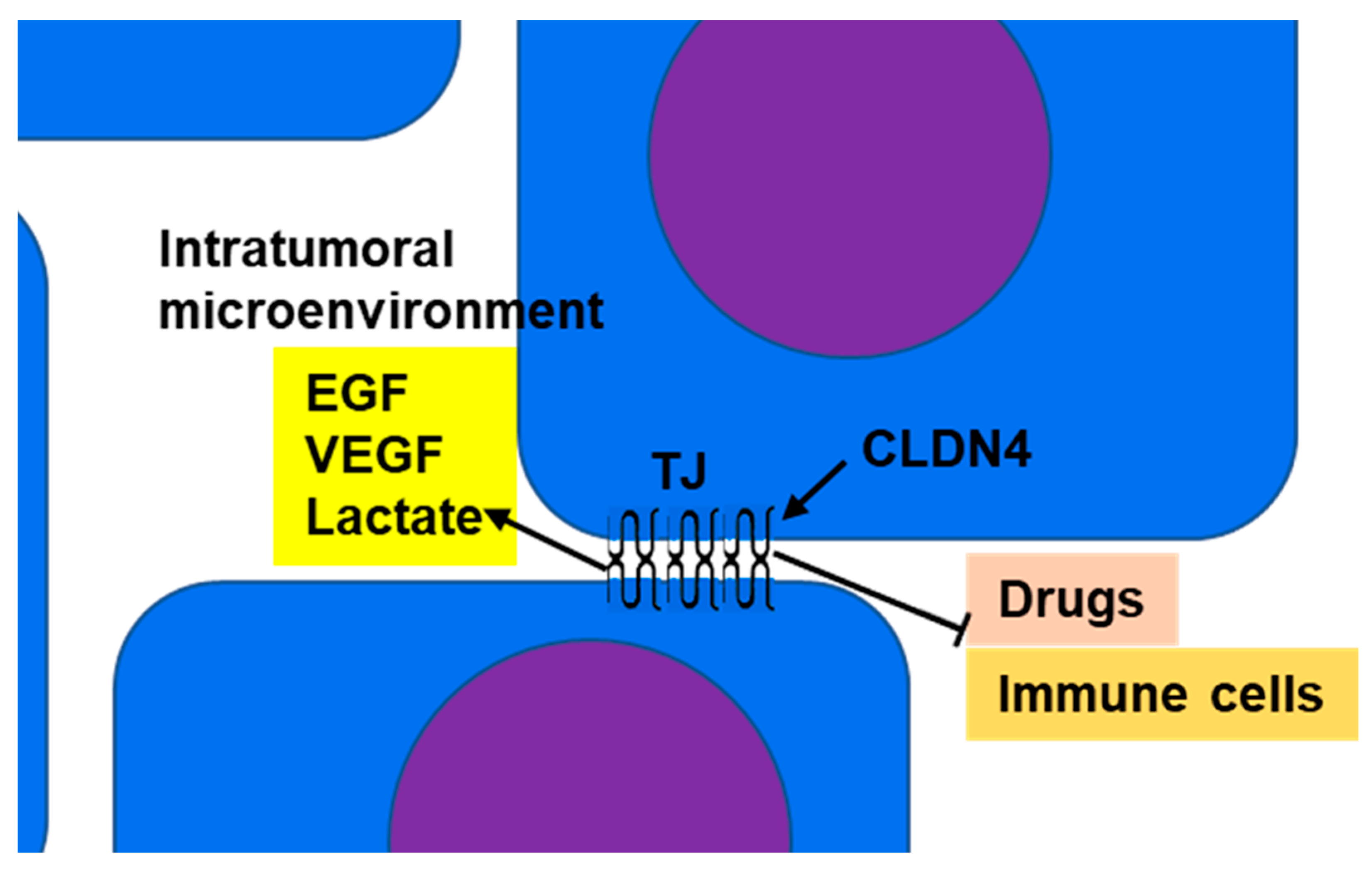
3.2. Barrier Function and Maintenance of Intratumoral Microenvironment
3.3. Apoptosis
3.4. Stemness and EMT
4. Non-TJ Functions of CLDN4
4.1. Non-TJ Plasma Membrane CLDN4
4.2. Cytoplasmic CLDN4
4.3. Function of Non-TJ CLDN4
4.3.1. Integrin β1 Activation
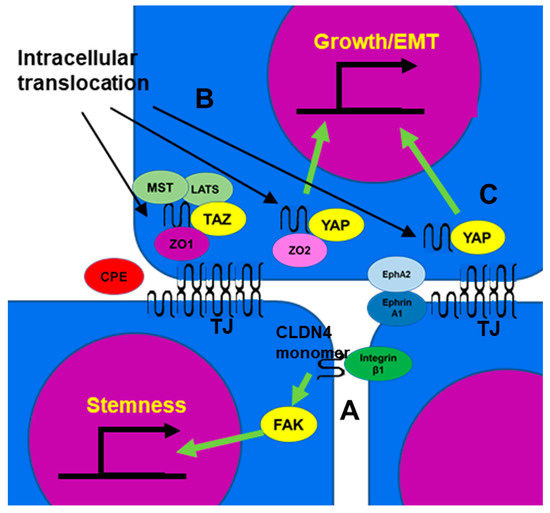
4.3.2. YAP Activation
4.3.3. Activation of AKT
5. Targeting CLDN4
5.1. Antibodies
5.2. Knockdown
5.3. CPE and C-Terminus Domain of CPE (C-CPE)
5.4. Peptide
5.5. Delivery of Anti-CLDN4 Drugs
References
- Otani, T.; Furuse, M. Tight Junction Structure and Function Revisited. Trends Cell Biol. 2020, 30, 805–817.
- Hashimoto, Y.; Fukasawa, M.; Kuniyasu, H.; Yagi, K.; Kondoh, M. Claudin-targeted drug development using anti-claudin monoclonal antibodies to treat hepatitis and cancer. Ann. N. Y. Acad. Sci. 2017, 1397, 5–16.
- Osanai, M.; Takasawa, A.; Murata, M.; Sawada, N. Claudins in cancer: Bench to bedside. Pflug. Arch. Eur. J. Physiol. 2017, 469, 55–67.
- Morin, P.J. Claudin proteins in human cancer: Promising new targets for diagnosis and therapy. Cancer Res. 2005, 65, 9603–9606.
- Neesse, A.; Griesmann, H.; Gress, T.M.; Michl, P. Claudin-4 as therapeutic target in cancer. Arch. Biochem. Biophys. 2012, 524, 64–70.
- Zhu, J.L.; Gao, P.; Wang, Z.N.; Song, Y.X.; Li, A.L.; Xu, Y.Y.; Wang, M.X.; Xu, H.M. Clinicopathological significance of claudin-4 in gastric carcinoma. World J. Surg. Oncol. 2013, 11, 150.
- Resnick, M.B.; Gavilanez, M.; Newton, E.; Konkin, T.; Bhattacharya, B.; Britt, D.E.; Sabo, E.; Moss, S.F. Claudin expression in gastric adenocarcinomas: A tissue microarray study with prognostic correlation. Hum. Pathol. 2005, 36, 886–892.
- Nishiguchi, Y.; Fujiwara-Tani, R.; Sasaki, T.; Luo, Y.; Ohmori, H.; Kishi, S.; Mori, S.; Goto, K.; Yasui, W.; Sho, M.; et al. Targeting claudin-4 enhances CDDP-chemosensitivity in gastric cancer. Oncotarget 2019, 10, 2189–2202.
- Nichols, L.S.; Ashfaq, R.; Iacobuzio-Donahue, C.A. Claudin 4 protein expression in primary and metastatic pancreatic cancer: Support for use as a therapeutic target. Am. J. Clin. Pathol. 2004, 121, 226–230.
- Michl, P.; Buchholz, M.; Rolke, M.; Kunsch, S.; Lohr, M.; McClane, B.; Tsukita, S.; Leder, G.; Adler, G.; Gress, T.M. Claudin-4: A new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology 2001, 121, 678–684.
- Sasaki, T.; Fujiwara-Tani, R.; Kishi, S.; Mori, S.; Luo, Y.; Ohmori, H.; Kawahara, I.; Goto, K.; Nishiguchi, Y.; Mori, T.; et al. Targeting claudin-4 enhances chemosensitivity of pancreatic ductal carcinomas. Cancer Med. 2019, 8, 6700–6708.
- De Oliveira, S.S.; de Oliveira, I.M.; De Souza, W.; Morgado-Díaz, J.A. Claudins upregulation in human colorectal cancer. FEBS Lett. 2005, 579, 6179–6185.
- Fujiwara-Tani, R.; Sasaki, T.; Luo, Y.; Goto, K.; Kawahara, I.; Nishiguchi, Y.; Kishi, S.; Mori, S.; Ohmori, H.; Kondoh, M.; et al. Anti-claudin-4 extracellular domain antibody enhances the antitumoral effects of chemotherapeutic and antibody drugs in colorectal cancer. Oncotarget 2018, 9, 37367–37378.
- Kolokytha, P.; Yiannou, P.; Keramopoulos, D.; Kolokythas, A.; Nonni, A.; Patsouris, E.; Pavlakis, K. Claudin-3 and claudin-4: Distinct prognostic significance in triple-negative and luminal breast cancer. Appl. Immunohistochem. Mol. Morphol. AIMM 2014, 22, 125–131.
- Ma, X.; Miao, H.; Jing, B.; Pan, Q.; Zhang, H.; Chen, Y.; Zhang, D.; Liang, Z.; Wen, Z.; Li, M. Claudin-4 controls the proliferation, apoptosis, migration and in vivo growth of MCF-7 breast cancer cells. Oncol. Rep. 2015, 34, 681–690.
- Luo, Y.; Kishi, S.; Sasaki, T.; Ohmori, H.; Fujiwara-Tani, R.; Mori, S.; Goto, K.; Nishiguchi, Y.; Mori, T.; Kawahara, I.; et al. Targeting claudin-4 enhances chemosensitivity in breast cancer. Cancer Sci. 2020, 111, 1840–1850.
- Naimi, A.; Zare, N.; Amjadi, E.; Soltan, M. High claudin-4 antigen expression in triple-negative breast cancer by the immunohistochemistry method. J. Res. Med. Sci. 2022, 27, 20.
- Nakashima, C.; Yamamoto, K.; Kishi, S.; Sasaki, T.; Ohmori, H.; Fujiwara-Tani, R.; Mori, S.; Kawahara, I.; Nishiguchi, Y.; Mori, T.; et al. Clostridium perfringens enterotoxin induces claudin-4 to activate YAP in oral squamous cell carcinomas. Oncotarget 2020, 11, 309–321.
- Rangel, L.B.; Agarwal, R.; D’Souza, T.; Pizer, E.S.; Alò, P.L.; Lancaster, W.D.; Gregoire, L.; Schwartz, D.R.; Cho, K.R.; Morin, P.J. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin. Cancer Res. 2003, 9, 2567–2575.
- Kuwada, M.; Chihara, Y.; Luo, Y.; Li, X.; Nishiguchi, Y.; Fujiwara, R.; Sasaki, T.; Fujii, K.; Ohmori, H.; Fujimoto, K.; et al. Pro-chemotherapeutic effects of antibody against extracellular domain of claudin-4 in bladder cancer. Cancer Lett. 2015, 369, 212–221.
- Maesaka, F.; Kuwada, M.; Horii, S.; Kishi, S.; Fujiwara-Tani, R.; Mori, S.; Fujii, K.; Mori, T.; Ohmori, H.; Owari, T.; et al. Hypomethylation of CLDN4 Gene Promoter Is Associated with Malignant Phenotype in Urinary Bladder Cancer. Int. J. Mol. Sci. 2022, 23, 6516.
- Piontek, A.; Eichner, M.; Zwanziger, D.; Beier, L.S.; Protze, J.; Walther, W.; Theurer, S.; Schmid, K.W.; Führer-Sakel, D.; Piontek, J.; et al. Targeting claudin-overexpressing thyroid and lung cancer by modified Clostridium perfringens enterotoxin. Mol. Oncol. 2020, 14, 261–276.
- Bunthot, S.; Obchoei, S.; Kraiklang, R.; Pirojkul, C.; Wongkham, S.; Wongkham, C. Overexpression of claudin-4 in cholangiocarcinoma tissues and its possible role in tumor metastasis. Asian Pac. J. Cancer Prev. 2012, 13, 71–76.
- Tzelepi, V.N.; Tsamandas, A.C.; Vlotinou, H.D.; Vagianos, C.E.; Scopa, C.D. Tight junctions in thyroid carcinogenesis: Diverse expression of claudin-1, claudin-4, claudin-7 and occludin in thyroid neoplasms. Mod. Pathol. 2008, 21, 22–30.
- Coutinho-Camillo, C.M.; Lourenço, S.V.; da Fonseca, F.P.; Soares, F.A. Claudin expression is dysregulated in prostate adenocarcinomas but does not correlate with main clinicopathological parameters. Pathology 2011, 43, 143–148.
- Sheehan, G.M.; Kallakury, B.V.; Sheehan, C.E.; Fisher, H.A.; Kaufman, R.P., Jr.; Ross, J.S. Loss of claudins-1 and -7 and expression of claudins-3 and -4 correlate with prognostic variables in prostatic adenocarcinomas. Hum. Pathol. 2007, 38, 564–569.
- Nebbioso, A.; Tambaro, F.P.; Dell’Aversana, C.; Altucci, L. Cancer epigenetics: Moving forward. PLoS Genet. 2018, 14, e1007362.
- Feinberg, A.P. The Key Role of Epigenetics in Human Disease Prevention and Mitigation. N. Engl. J. Med. 2018, 378, 1323–1334.
- Kanwal, R.; Gupta, K.; Gupta, S. Cancer epigenetics: An introduction. Methods Mol. Biol. 2015, 1238, 3–25.
- Kwon, M.J.; Kim, S.S.; Choi, Y.L.; Jung, H.S.; Balch, C.; Kim, S.H.; Song, Y.S.; Marquez, V.E.; Nephew, K.P.; Shin, Y.K. Derepression of CLDN3 and CLDN4 during ovarian tumorigenesis is associated with loss of repressive histone modifications. Carcinogenesis 2010, 31, 974–983.
- Boireau, S.; Buchert, M.; Samuel, M.S.; Pannequin, J.; Ryan, J.L.; Choquet, A.; Chapuis, H.; Rebillard, X.; Avances, C.; Ernst, M.; et al. DNA-methylation-dependent alterations of claudin-4 expression in human bladder carcinoma. Carcinogenesis 2007, 28, 246–258.
- Hahn-Stromberg, V.; Askari, S.; Ahmad, A.; Befekadu, R.; Nilsson, T.K. Expression of claudin 1, claudin 4, and claudin 7 in colorectal cancer and its relation with CLDN DNA methylation patterns. Tumor Biol. 2017, 39, 1010428317697569.
- Kwon, M.J.; Kim, S.H.; Jeong, H.M.; Jung, H.S.; Kim, S.S.; Lee, J.E.; Gye, M.C.; Erkin, O.C.; Koh, S.S.; Choi, Y.L.; et al. Claudin-4 overexpression is associated with epigenetic derepression in gastric carcinoma. Lab. Investig. 2011, 91, 1652–1667.
- Ma, M.C.; Qian, H.; Ghassemi, F.; Zhao, P.; Xia, Y. Oxygen-sensitive δ-opioid receptor-regulated survival and death signals: Novel insights into neuronal preconditioning and protection. J. Biol. Chem. 2005, 280, 16208–16218.
- Litkouhi, B.; Kwong, J.; Lo, C.M.; Smedley, J.G., 3rd; McClane, B.A.; Aponte, M.; Gao, Z.; Sarno, J.L.; Hinners, J.; Welch, W.R.; et al. Claudin-4 overexpression in epithelial ovarian cancer is associated with hypomethylation and is a potential target for modulation of tight junction barrier function using a C-terminal fragment of Clostridium perfringens enterotoxin. Neoplasia 2007, 9, 304–314.
- Hasegawa, K.; Wakino, S.; Simic, P.; Sakamaki, Y.; Minakuchi, H.; Fujimura, K.; Hosoya, K.; Komatsu, M.; Kaneko, Y.; Kanda, T.; et al. Renal tubular Sirt1 attenuates diabetic albuminuria by epigenetically suppressing Claudin-1 overexpression in podocytes. Nat. Med. 2013, 19, 1496–1504.
- Di Cello, F.; Cope, L.; Li, H.; Jeschke, J.; Wang, W.; Baylin, S.B.; Zahnow, C.A. Methylation of the claudin 1 promoter is associated with loss of expression in estrogen receptor positive breast cancer. PLoS ONE 2013, 8, e68630.
- Hichino, A.; Okamoto, M.; Taga, S.; Akizuki, R.; Endo, S.; Matsunaga, T.; Ikari, A. Down-regulation of Claudin-2 Expression and Proliferation by Epigenetic Inhibitors in Human Lung Adenocarcinoma A549 Cells. J. Biol. Chem. 2017, 292, 2411–2421.
- Zhang, Z.; Yu, W.; Chen, S.; Chen, Y.; Chen, L.; Zhang, S. Methylation of the claudin3 promoter predicts the prognosis of advanced gastric adenocarcinoma. Oncol. Rep. 2018, 40, 49–60.
- Honda, H.; Pazin, M.J.; D’Souza, T.; Ji, H.; Morin, P.J. Regulation of the CLDN3 gene in ovarian cancer cells. Cancer Biol. Ther. 2007, 6, 1733–1742.
- Nishikiori, N.; Sawada, N.; Ohguro, H. Prevention of murine experimental corneal trauma by epigenetic events regulating claudin 6 and claudin 9. Jpn. J. Ophthalmol. 2008, 52, 195–203.
- Kudinov, A.E.; Deneka, A.; Nikonova, A.S.; Beck, T.N.; Ahn, Y.H.; Liu, X.; Martinez, C.F.; Schultz, F.A.; Reynolds, S.; Yang, D.H.; et al. Musashi-2 (MSI2) supports TGF-β signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis. Proc. Natl. Acad. Sci. USA 2016, 113, 6955–6960.
- Li, J.; Zhou, C.; Ni, S.; Wang, S.; Ni, C.; Yang, P.; Ye, M. Methylated claudin-11 associated with metastasis and poor survival of colorectal cancer. Oncotarget 2017, 8, 96249–96262.
- Jie, Y.; Ye, L.; Chen, H.; Yu, X.; Cai, L.; He, W.; Fu, Y. ELFN1-AS1 accelerates cell proliferation, invasion and migration via regulating miR-497-3p/CLDN4 axis in ovarian cancer. Bioengineered 2020, 11, 872–882.
- Satake, S.; Semba, S.; Matsuda, Y.; Usami, Y.; Chiba, H.; Sawada, N.; Kasuga, M.; Yokozaki, H. Cdx2 transcription factor regulates claudin-3 and claudin-4 expression during intestinal differentiation of gastric carcinoma. Pathol. Int. 2008, 58, 156–163.
- Kodera, Y.; Kohno, T.; Konno, T.; Arai, W.; Tsujiwaki, M.; Shindo, Y.; Chiba, H.; Miyakawa, M.; Tanaka, H.; Sakuma, Y.; et al. HMGB1 enhances epithelial permeability via p63/TGF-β signaling in lung and terminal bronchial epithelial cells. Tissue Barriers 2020, 8, 1805997.
- Nur Husna, S.M.; Md Shukri, N.; Tuan Sharif, S.E.; Tan, H.T.T.; Mohd Ashari, N.S.; Wong, K.K. IL-4/IL-13 Axis in Allergic Rhinitis: Elevated Serum Cytokines Levels and Inverse Association with Tight Junction Molecules Expression. Front. Mol. Biosci. 2022, 9, 819772.
- Yang, Y.; Cheon, S.; Jung, M.K.; Song, S.B.; Kim, D.; Kim, H.J.; Park, H.; Bang, S.I.; Cho, D. Interleukin-18 enhances breast cancer cell migration via down-regulation of claudin-12 and induction of the p38 MAPK pathway. Biochem. Biophys. Res. Commun. 2015, 459, 379–386.
- Rachakonda, G.; Vu, T.; Jin, L.; Samanta, D.; Datta, P.K. Role of TGF-β-induced Claudin-4 expression through c-Jun signaling in non-small cell lung cancer. Cell. Signal. 2016, 28, 1537–1544.
- Marincola Smith, P.; Choksi, Y.A.; Markham, N.O.; Hanna, D.N.; Zi, J.; Weaver, C.J.; Hamaamen, J.A.; Lewis, K.B.; Yang, J.; Liu, Q.; et al. Colon epithelial cell TGFβ signaling modulates the expression of tight junction proteins and barrier function in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G936–G957.
- Yan, T.; Tan, Y.; Deng, G.; Sun, Z.; Liu, B.; Wang, Y.; Yuan, F.; Sun, Q.; Hu, P.; Gao, L.; et al. TGF-β induces GBM mesenchymal transition through upregulation of CLDN4 and nuclear translocation to activate TNF-α/NF-κB signal pathway. Cell Death Dis. 2022, 13, 339.
- Kojima, T.; Kyuno, D.; Sawada, N. Targeting claudin-4 in human pancreatic cancer. Expert Opin. Ther. Targets 2012, 16, 881–887.
- Väre, P.; Soini, Y. Twist is inversely associated with claudins in germ cell tumors of the testis. APMIS 2010, 118, 640–647.
- Kim, B.; Breton, S. The MAPK/ERK-Signaling Pathway Regulates the Expression and Distribution of Tight Junction Proteins in the Mouse Proximal Epididymis. Biol. Reprod. 2016, 94, 22.
- Ishii, Y.; Saeki, K.; Liu, M.; Sasaki, F.; Koga, T.; Kitajima, K.; Meno, C.; Okuno, T.; Yokomizo, T. Leukotriene B4 receptor type 2 (BLT2) enhances skin barrier function by regulating tight junction proteins. FASEB J. 2016, 30, 933–947.
- Liu, H.; Zhang, Z.; Zhou, S.; Liu, X.; Li, G.; Song, B.; Xu, W. Claudin-1/4 as directly target gene of HIF-1α can feedback regulating HIF-1α by PI3K-AKT-mTOR and impact the proliferation of esophageal squamous cell though Rho GTPase and p-JNK pathway. Cancer Gene Ther. 2022, 29, 665–682.
- Batsaikhan, B.E.; Yoshikawa, K.; Kurita, N.; Iwata, T.; Takasu, C.; Kashihara, H.; Shimada, M. Cyclopamine decreased the expression of Sonic Hedgehog and its downstream genes in colon cancer stem cells. Anticancer Res. 2014, 34, 6339–6344.
- Yamada, G.; Murata, M.; Takasawa, A.; Nojima, M.; Mori, Y.; Sawada, N.; Takahashi, H. Increased expressions of claudin 4 and 7 in atypical adenomatous hyperplasia and adenocarcinoma of the lung. Med. Mol. Morphol. 2016, 49, 163–169.
- Seckin, Y.; Arici, S.; Harputluoglu, M.; Yonem, O.; Yilmaz, A.; Ozer, H.; Karincaoglu, M.; Demirel, U. Expression of claudin-4 and beta-catenin in gastric premalignant lesions. Acta Gastroenterol. Belg. 2009, 72, 407–412.
- Fujiwara-Tani, R.; Fujii, K.; Mori, S.; Kishi, S.; Sasaki, T.; Ohmori, H.; Nakashima, C.; Kawahara, I.; Nishiguchi, Y.; Mori, T.; et al. Role of Clostridium perfringens Enterotoxin on YAP Activation in Colonic Sessile Serrated Adenoma/Polyps with Dysplasia. Int. J. Mol. Sci. 2020, 21, 3840.
- Pan, X.Y.; Wang, B.; Che, Y.C.; Weng, Z.P.; Dai, H.Y.; Peng, W. Expression of claudin-3 and claudin-4 in normal, hyperplastic, and malignant endometrial tissue. Int. J. Gynecol. Cancer 2007, 17, 233–241.
- Sobel, G.; Páska, C.; Szabó, I.; Kiss, A.; Kádár, A.; Schaff, Z. Increased expression of claudins in cervical squamous intraepithelial neoplasia and invasive carcinoma. Hum. Pathol. 2005, 36, 162–169.
- Zhu, Y.; Brännström, M.; Janson, P.O.; Sundfeldt, K. Differences in expression patterns of the tight junction proteins, claudin 1, 3, 4 and 5, in human ovarian surface epithelium as compared to epithelia in inclusion cysts and epithelial ovarian tumours. Int. J. Cancer 2006, 118, 1884–1891.
- Turksen, K.; Troy, T.C. Junctions gone bad: Claudins and loss of the barrier in cancer. Biochim. Biophys. Acta 2011, 1816, 73–79.
- Escudero-Esparza, A.; Jiang, W.G.; Martin, T.A. The Claudin family and its role in cancer and metastasis. Front. Biosci. 2011, 16, 1069–1083.
- Pade, V.; Stavchansky, S. Estimation of the relative contribution of the transcellular and paracellular pathway to the transport of passively absorbed drugs in the Caco-2 cell culture model. Pharm. Res. 1997, 14, 1210–1215.
- Hicks, D.A.; Galimanis, C.E.; Webb, P.G.; Spillman, M.A.; Behbakht, K.; Neville, M.C.; Baumgartner, H.K. Claudin-4 activity in ovarian tumor cell apoptosis resistance and migration. BMC Cancer 2016, 16, 788.
- Pao, H.P.; Liao, W.I.; Tang, S.E.; Wu, S.Y.; Huang, K.L.; Chu, S.J. Suppression of Endoplasmic Reticulum Stress by 4-PBA Protects Against Hyperoxia-Induced Acute Lung Injury via Up-Regulating Claudin-4 Expression. Front. Immunol. 2021, 12, 674316.
- Singh, A.B.; Sharma, A.; Dhawan, P. Claudin-1 expression confers resistance to anoikis in colon cancer cells in a Src-dependent manner. Carcinogenesis 2012, 33, 2538–2547.
- Huang, J.; Zhang, L.; He, C.; Qu, Y.; Li, J.; Zhang, J.; Du, T.; Chen, X.; Yu, Y.; Liu, B.; et al. Claudin-1 enhances tumor proliferation and metastasis by regulating cell anoikis in gastric cancer. Oncotarget 2015, 6, 1652–1665.
- Osanai, M.; Murata, M.; Chiba, H.; Kojima, T.; Sawada, N. Epigenetic silencing of claudin-6 promotes anchorage-independent growth of breast carcinoma cells. Cancer Sci. 2007, 98, 1557–1562.
- Najafi, M.; Mortezaee, K.; Majidpoor, J. Cancer stem cell (CSC) resistance drivers. Life Sci. 2019, 234, 116781.
- Ayob, A.Z.; Ramasamy, T.S. Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018, 25, 20.
- Kwon, M.J. Emerging roles of claudins in human cancer. Int. J. Mol. Sci. 2013, 14, 18148–18180.
- Kyuno, D.; Yamaguchi, H.; Ito, T.; Kono, T.; Kimura, Y.; Imamura, M.; Konno, T.; Hirata, K.; Sawada, N.; Kojima, T. Targeting tight junctions during epithelial to mesenchymal transition in human pancreatic cancer. World J. Gastroenterol. 2014, 20, 10813–10824.
- Papageorgis, P.; Lambert, A.W.; Ozturk, S.; Gao, F.; Pan, H.; Manne, U.; Alekseyev, Y.O.; Thiagalingam, A.; Abdolmaleky, H.M.; Lenburg, M.; et al. Smad signaling is required to maintain epigenetic silencing during breast cancer progression. Cancer Res. 2010, 70, 968–978.
- Sato, M.; Matsumoto, M.; Saiki, Y.; Alam, M.; Nishizawa, H.; Rokugo, M.; Brydun, A.; Yamada, S.; Kaneko, M.K.; Funayama, R.; et al. BACH1 Promotes Pancreatic Cancer Metastasis by Repressing Epithelial Genes and Enhancing Epithelial-Mesenchymal Transition. Cancer Res. 2020, 80, 1279–1292.
- Shrestha, A.; Uzal, F.A.; McClane, B.A. The interaction of Clostridium perfringens enterotoxin with receptor claudins. Anaerobe 2016, 41, 18–26.
- Gardelli, C.; Russo, L.; Cipolla, L.; Moro, M.; Andriani, F.; Rondinone, O.; Nicotra, F.; Sozzi, G.; Bertolini, G.; Roz, L. Differential glycosylation of collagen modulates lung cancer stem cell subsets through β1 integrin-mediated interactions. Cancer Sci. 2021, 112, 217–230.
- Moon, J.H.; Rho, Y.S.; Lee, S.H.; Koo, B.S.; Lee, H.J.; Do, S.I.; Cho, J.H.; Eun, Y.G.; Park, M.W.; Shin, H.A.; et al. Role of integrin β1 as a biomarker of stemness in head and neck squamous cell carcinoma. Oral Oncol. 2019, 96, 34–41.
- Lu, Z.; Kim, D.H.; Fan, J.; Lu, Q.; Verbanac, K.; Ding, L.; Renegar, R.; Chen, Y.H. A non-tight junction function of claudin-7-Interaction with integrin signaling in suppressing lung cancer cell proliferation and detachment. Mol. Cancer 2015, 14, 120.
- Kim, D.H.; Lu, Q.; Chen, Y.H. Claudin-7 modulates cell-matrix adhesion that controls cell migration, invasion and attachment of human HCC827 lung cancer cells. Oncol. Lett. 2019, 17, 2890–2896.
- Owari, T.; Sasaki, T.; Fujii, K.; Fujiwara-Tani, R.; Kishi, S.; Mori, S.; Mori, T.; Goto, K.; Kawahara, I.; Nakai, Y.; et al. Role of Nuclear Claudin-4 in Renal Cell Carcinoma. Int. J. Mol. Sci. 2020, 21, 8340.
- Yu, S.; Zhang, Y.; Li, Q.; Zhang, Z.; Zhao, G.; Xu, J. CLDN6 promotes tumor progression through the YAP1-snail1 axis in gastric cancer. Cell Death Dis. 2019, 10, 949.
- Kong, F.E.; Li, G.M.; Tang, Y.Q.; Xi, S.Y.; Loong, J.H.C.; Li, M.M.; Li, H.L.; Cheng, W.; Zhu, W.J.; Mo, J.Q.; et al. Targeting tumor lineage plasticity in hepatocellular carcinoma using an anti-CLDN6 antibody-drug conjugate. Sci. Transl. Med. 2021, 13, eabb6282.
- Zhou, B.; Flodby, P.; Luo, J.; Castillo, D.R.; Liu, Y.; Yu, F.X.; McConnell, A.; Varghese, B.; Li, G.; Chimge, N.O.; et al. Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J. Clin. Investig. 2018, 128, 970–984.
- Kohmoto, T.; Masuda, K.; Shoda, K.; Takahashi, R.; Ujiro, S.; Tange, S.; Ichikawa, D.; Otsuji, E.; Imoto, I. Claudin-6 is a single prognostic marker and functions as a tumor-promoting gene in a subgroup of intestinal type gastric cancer. Gastric Cancer 2020, 23, 403–417.
- Hao, S.; Yang, C.; Song, P.; Shi, H.; Zou, Y.; Chen, M.; Wu, X.; Yin, Y.; Yu, Z.; Zhu, W.; et al. CLDN4 promotes growth of acute myeloid leukemia cells via regulating AKT and ERK1/2 signaling. Biochem. Biophys. Res. Commun. 2022, 619, 137–143.
- Wang, P.; Liu, T.; Zhao, Z.; Wang, Z.; Liu, S.; Yang, X. SPTBN2 regulated by miR-424-5p promotes endometrial cancer progression via CLDN4/PI3K/AKT axis. Cell Death Discov. 2021, 7, 382.
- Luo, J.; Wang, H.; Chen, H.; Gan, G.; Zheng, Y. CLDN4 silencing promotes proliferation and reduces chemotherapy sensitivity of gastric cancer cells through activation of the PI3K/Akt signalling pathway. Exp. Physiol. 2020, 105, 979–988.
- Shang, X.; Lin, X.; Howell, S.B. Claudin-4 controls the receptor tyrosine kinase EphA2 pro-oncogenic switch through β-catenin. Cell Commun. Signal. 2014, 12, 59.
- Hashimoto, Y.; Okada, Y.; Shirakura, K.; Tachibana, K.; Sawada, M.; Yagi, K.; Doi, T.; Kondoh, M. Anti-Claudin Antibodies as a Concept for Development of Claudin-Directed Drugs. J. Pharmacol. Exp. Ther. 2019, 368, 179–186.
- Kato-Nakano, M.; Suzuki, M.; Kawamoto, S.; Furuya, A.; Ohta, S.; Nakamura, K.; Ando, H. Characterization and evaluation of the antitumour activity of a dual-targeting monoclonal antibody against claudin-3 and claudin-4. Anticancer Res. 2010, 30, 4555–4562.
- Li, X.; Iida, M.; Tada, M.; Watari, A.; Kawahigashi, Y.; Kimura, Y.; Yamashita, T.; Ishii-Watabe, A.; Uno, T.; Fukasawa, M.; et al. Development of an anti-claudin-3 and -4 bispecific monoclonal antibody for cancer diagnosis and therapy. J. Pharmacol. Exp. Ther. 2014, 351, 206–213.
- Suzuki, M.; Kato-Nakano, M.; Kawamoto, S.; Furuya, A.; Abe, Y.; Misaka, H.; Kimoto, N.; Nakamura, K.; Ohta, S.; Ando, H. Therapeutic antitumor efficacy of monoclonal antibody against Claudin-4 for pancreatic and ovarian cancers. Cancer Sci. 2009, 100, 1623–1630.
- Mitchell, L.A.; Koval, M. Specificity of interaction between clostridium perfringens enterotoxin and claudin-family tight junction proteins. Toxins 2010, 2, 1595–1611.
- Romanov, V.; Whyard, T.C.; Waltzer, W.C.; Gabig, T.G. A claudin 3 and claudin 4-targeted Clostridium perfringens protoxin is selectively cytotoxic to PSA-producing prostate cancer cells. Cancer Lett. 2014, 351, 260–264.
- Landers, K.A.; Samaratunga, H.; Teng, L.; Buck, M.; Burger, M.J.; Scells, B.; Lavin, M.F.; Gardiner, R.A. Identification of claudin-4 as a marker highly overexpressed in both primary and metastatic prostate cancer. Br. J. Cancer 2008, 99, 491–501.
- Liang, Z.Y.; Kang, X.; Chen, H.; Wang, M.; Guan, W.X. Effect of Clostridium perfringens enterotoxin on gastric cancer cells SGC7901 which highly expressed claudin-4 protein. World J. Gastrointestig. Oncol. 2017, 9, 153–159.
- Tanaka, S.; Aoyama, T.; Ogawa, M.; Takasawa, A.; Murata, M.; Osanai, M.; Saito, T.; Sawada, N. Cytotoxicity of Clostridium perfringens enterotoxin depends on the conditions of claudin-4 in ovarian carcinoma cells. Exp. Cell Res. 2018, 371, 278–286.
- English, D.P.; Santin, A.D. Claudins overexpression in ovarian cancer: Potential targets for Clostridium Perfringens Enterotoxin (CPE) based diagnosis and therapy. Int. J. Mol. Sci. 2013, 14, 10412–10437.




