
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Deirdre Hanrahan-Tan | -- | 1413 | 2023-05-22 08:41:53 | | | |
| 2 | Sirius Huang | Meta information modification | 1413 | 2023-05-22 09:11:16 | | |
Video Upload Options
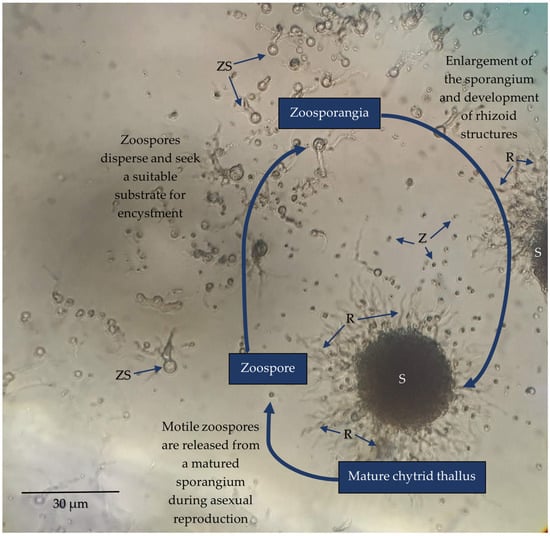
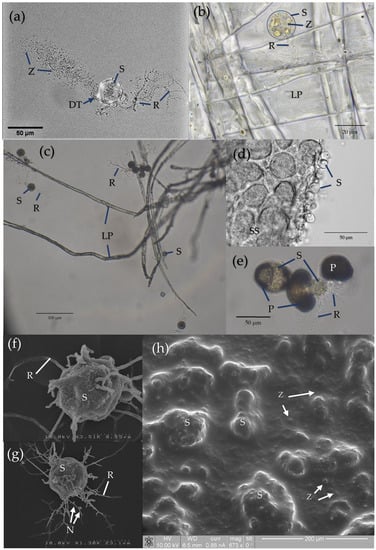
Chytridiomycota (zoosporic true fungi) have a consistent presence in soils and have been frequently identified within many diverse terrestrial environments. However, Chytridiomycota and other early-diverging fungi have low representation in whole-genome sequencing databases compared to Dikarya. New molecular techniques have provided insights into the diversity and abundance of chytrids in soils and the changes in their populations both spatially and temporally. Chytrids complete their life cycle within rapidly changing soil environments where they may be more common within micropores due to protection from predation, desiccation, and extreme temperatures. Reproductive and morphological changes occur in response to environmental changes including pH, fluctuating nutrient concentrations, and metals at levels above toxic thresholds. Rhizoids share some features of hyphae, including the spatial regulation of branching and the ability to attach, adapt to, and proliferate in different substrates, albeit on a microscale. Soil chytrids provide a pool of novel enzymes and proteins which enable a range of lifestyles as saprotrophs or parasites, but also can be utilised as alternative tools with some biotechnological applications. Thus, 3D live-cell imaging and micromodels such as MicroCT may provide insight into zoospore functions and rhizoid plasticity, respectively, in response to various conditions. A combination of classical techniques of soil chytrid baiting with simultaneous molecular and ecological data will provide insights into temporal population changes in response to environmental change. The authors emphasise the need to review and improve DNA-based methodologies for identifying and quantifying chytrids within the soil microbiome to expand our knowledge of their taxonomy, abundance, diversity, and functionality within soil environments.
References
- Sparrow, F.K. Aquatic Phycomycetes, 2nd ed.; University of Michigan Press: Ann Arbor, MI, USA, 1960.
- Willoughby, L.G. The Ecology of some lower fungi at Esthwaite water. Trans. Br. Mycol. Soc. 1961, 44, 305–332, IN1–IN2.
- Kagami, M.; de Bruin, A.; Ibelings, B.W.; Van Donk, E. Parasitic chytrids: Their effects on phytoplankton communities and food-web dynamics. Hydrobiologia 2007, 578, 113–129.
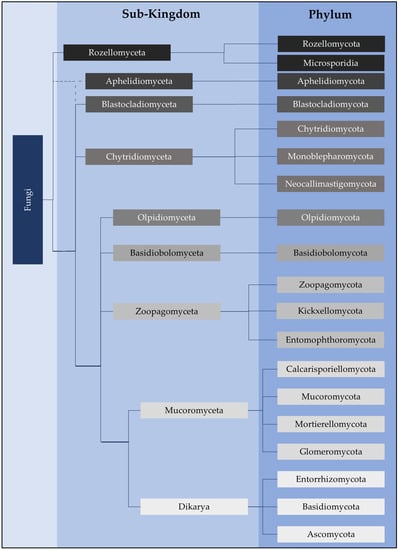
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal evolution: Diversity, taxonomy and phylogeny of the Fungi. Biol. Rev. Camb. Philos. Soc. 2019, 94, 2101–2137.
- Tedersoo, L.; Bahram, M.; Puusepp, R.; Nilsson, R.H.; James, T. Novel soil-inhabiting clades fill gaps in the fungal tree of life. Microbiome 2017, 5, 42.
- Li, Y.; Steenwyk, J.L.; Chang, Y.; Wang, Y.; James, T.Y.; Stajich, J.E.; Spatafora, J.W.; Groenewald, M.; Dunn, C.W.; Hittinger, C.T.; et al. A genome-scale phylogeny of the kingdom Fungi. Curr. Biol. 2021, 31, 1653–1665.e5.
- James, T.Y.; Stajich, J.E.; Hittinger, C.T.; Rokas, A. Toward a Fully Resolved Fungal Tree of Life. Annu. Rev. Microbiol. 2020, 74, 291–313.
- Baldauf, S.L. The Deep Roots of Eukaryotes. Sci. Am. Assoc. Adv. Sci. 2003, 300, 1703–1706.
- Baldauf, S.L. An overview of the phylogeny and diversity of eukaryotes. J. Syst. Evol. 2008, 46, 263–273.
- Ruggiero, M.A.; Gordon, D.P.; Orrell, T.M.; Bailly, N.; Bourgoin, T.; Brusca, R.C.; Cavalier-Smith, T.; Guiry, M.D.; Kirk, P.M. A higher level classification of all living organisms. PLoS ONE 2015, 10, e0119248.
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Div. 2018, 90, 135–159.
- Powell, M.J.; Letcher, P.M. Chytridiomycota, Monoblepharidomycota, and Neocallimastigomycota. In The Mycota, 2nd ed.; Esser, K., McLaughlin, D.J., Spatafora, J.W., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 7, pp. 141–175.
- Strassert, J.F.H.; Wurzbacher, C.; Hervé, V.; Antany, T.; Brune, A.; Radek, R. Long rDNA amplicon sequencing of insect-infecting nephridiophagids reveals their affiliation to the Chytridiomycota and a potential to switch between hosts. Sci. Rep. 2021, 11, 396.
- Hibbett, D.S.; Binder, M.; Bischoff, J.F.; Blackwell, M.; Cannon, P.F.; Eriksson, O.E.; Huhndorf, S.; James, T.; Kirk, P.M.; Lücking, R.; et al. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 2007, 111, 509–547.
- James, T.Y.; Letcher, P.M.; Longcore, J.E.; Mozley-Standridge, S.E.; Porter, D.; Powell, M.J.; Griffith, G.W.; Vilgalys, R. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 2006, 98, 860–871.
- Powell, M.J. Chytridiomycota. In Handbook of the Protists, 2nd ed.; Archibald, J., Simpson, A., Slamovits, C., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1523–1558.
- Porter, T.M.; Martin, W.; James, T.Y.; Longcore, J.E.; Gleason, F.H.; Adler, P.H.; Letcher, P.M.; Vilgalys, R. Molecular phylogeny of the Blastocladiomycota (Fungi) based on nuclear ribosomal DNA. Fungal Biol. 2011, 115, 381–392.
- James, T.Y.; Kauff, F.; Schoch, C.L.; Matheny, P.B.; Hofstetter, V.; Cox, C.J.; Celio, G.; Gueidan, C.; Fraker, E.; Miadlikowska, J. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 2006, 443, 818–822.
- Gleason, F.H.; Crawford, J.W.; Neuhauser, S.; Henderson, L.E.; Lilje, O. Resource seeking strategies of zoosporic true fungi in heterogeneous soil habitats at the microscale level. Soil Biol. Biochem. 2012, 45, 79–88.
- Held, A.A. Attraction and attachment of zoospores of the parasitic chytrid Rozella allomycis in response to host-dependent factors. Arch. Microbiol. 1974, 95, 97–114.
- Mitchell, R.T.; Deacon, J.W. Selective accumulation of zoospores of chytridiomycetes and oomycetes on cellulose and chitin. Trans. Br. Mycol. Soc. 1986, 86, 219–223.
- Gleason, F.H.; Lilje, O. Structure and function of fungal zoospores: Ecological implications. Fungal Ecol. 2009, 2, 53–59.
- Weete, J.D.; Laseter, J.L. Distribution of sterols in the fungi I. Fungal spores. Lipids 1974, 9, 575–581.
- Beakes, G.W.; Canter, H.M.; Jaworski, G.H.M. Comparative ultrastructural ontogeny of zoosporangia of Zygorhizidium affluens and Z. planktonicum, chytrid parasites of the diatom Asterionella formosa. Mycol. Res. 1992, 96, 1047–1059.
- Barr, D.J.S. Phlyctochytrium reinboldtae (Chytridiales): Morphology and physiology. Can. J. Bot. 1970, 48, 479–484.
- Scheele, B.C.; Skerratt, L.F.; Grogan, L.F.; Hunter, D.A.; Clemann, N.; McFadden, M.; Newell, D.; Hoskin, C.J.; Gillespie, G.R.; Heard, G.W. After the epidemic: Ongoing declines, stabilizations and recoveries in amphibians afflicted by chytridiomycosis. Biol. Conserv. 2017, 206, 37–46.
- Scheele, B.C.; Pasmans, F.; Skerratt, L.F.; Berger, L.; Martel, A.; Beukema, W.; Acevedo, A.A.; Burrowes, P.A.; Carvalho, T.; Catenazzi, A.; et al. Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science 2019, 363, 1459–1463.
- Tessa, G.; Sotgiu, G.; Bovero, S.; Angelini, C.; Favelli, M.; Gazzaniga, E.; Giacoma, C.; Garner, T.W.J. Cryptic but direct costs of an epidemic caused by Batrachochytrium dendrobatidis in the endangered Sardinian newt Euproctus platycephalus (Amphibia, Caudata). Amphibia-Reptilia 2023, 44, 83–94.
- O’Hanlon, S.J.; Rieux, A.; Farrer, R.A.; Rosa, G.M.; Waldman, B.; Bataille, A.; Kosch, T.A.; Murray, K.; Brankovics, B.; Fumagalli, M.; et al. A 20th Century Out-of-Asia Origin of a panzootic threat to global amphibian biodiversity. Science 2018, 360, 621–627.
- Weldon, C.; Channing, A.; Misinzo, G.; Cunningham, A.A. Disease driven extinction in the wild of the Kihansi spray toad, Nectophrynoides Asperginis. Afr. J. Herpetol. 2020, 69, 151–164.
- Klawonn, I.; Van den Wyngaert, S.; Iversen, M.H.; Walles, T.J.W.; Flintrop, C.M.; Cisternas-Novoa, C.; Nejstgaard, J.C.; Kagami, M.; Grossart, H.-P. Fungal parasitism on diatoms alters formation and bio–physical properties of sinking aggregates. Commun. Biol. 2023, 6, 206.
- Kagami, M.; Gurung, T.; Yoshida, T.; Urabe, J. To sink or to be lysed? Contrasting fate of two large phytoplankton species in Lake Biwa. Limnol. Oceanogr. 2006, 51, 2775–2786.
- Gerphagnon, M.; Colombet, J.; Latour, D.; Sime-Ngando, T. Spatial and temporal changes of parasitic chytrids of cyanobacteria. Sci. Rep. 2017, 7, 6056.
- Ibelings, B.W.; Gsell, A.S.; Mooij, W.M.; Van Donk, E.; Van Den Wyngaert, S.; De Senerpont Domis, L.N. Chytrid infections and diatom spring blooms: Paradoxical effects of climate warming on fungal epidemics in lakes. Freshw. Biol. 2011, 56, 754–766.
- Van den Wyngaert, S.; Ganzert, L.; Seto, K.; Rojas-Jimenez, K.; Agha, R.; Berger, S.A.; Woodhouse, J.; Padisak, J.; Wurzbacher, C.; Kagami, M.; et al. Seasonality of parasitic and saprotrophic zoosporic fungi: Linking sequence data to ecological traits. ISME J. 2022, 16, 2242–2254.
- Gsell, A.S.; Wolinska, J.; Preuß, K.; Teurlincx, S.; Özkundakci, D.; Hilt, S.; van Donk, E.; Ibelings, B.W.; Adrian, R. Long-term trends and seasonal variation in host density, temperature, and nutrients differentially affect chytrid fungi parasitising lake phytoplankton. Freshw. Biol. 2022, 67, 1532–1542.
- Lepelletier, F.; Karpov, S.A.; Alacid, E.; Le Panse, S.; Bigeard, E.; Garcés, E.; Jeanthon, C.; Guillou, L. Dinomyces arenysensis gen. et sp. nov. (Rhizophydiales, Dinomycetaceae fam. nov.), a Chytrid Infecting Marine Dinoflagellates. Protist 2014, 165, 230–244.
- Hanrahan-Tan, D.G.; Henderson, L.; Kertesz, M.A.; Lilje, O. The Effects of Nitrogen and Phosphorus on Colony Growth and Zoospore Characteristics of Soil Chytridiomycota. J. Fungi 2022, 8, 341.
- Henderson, L.; Marano, A.V.; Truszewski, E.; Gleason, F.H.; Lilje, O. Copper (II), lead (ll) and zinc (ll) reduce the rate of attachment in three zoosporic true fungi from soils of NSW, Australia. Nova Hedwig. 2019, 108, 435–447.
- Henderson, L.E.; Lilje, E.; Robinson, K.; Gleason, F.H.; Lilje, O. Effects of Toxic Metals on Chytrids, Fungal-Like Organisms, and Higher Fungi. In The Fungal Community: Its organisation and Role in the Ecosystem, 4th ed.; Dighton, J., White, J.F., Eds.; CRC Press: Boca Raton, FL, USA, 2017; Volume 32, pp. 487–512.





