Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Carlo Airola | -- | 5200 | 2023-05-21 21:17:10 | | | |
| 2 | Rita Xu | Meta information modification | 5200 | 2023-05-22 03:54:09 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Airola, C.; Severino, A.; Porcari, S.; Fusco, W.; Mullish, B.H.; Gasbarrini, A.; Cammarota, G.; Ponziani, F.R.; Ianiro, G. Future Modulation of Gut Microbiota. Encyclopedia. Available online: https://encyclopedia.pub/entry/44613 (accessed on 28 February 2026).
Airola C, Severino A, Porcari S, Fusco W, Mullish BH, Gasbarrini A, et al. Future Modulation of Gut Microbiota. Encyclopedia. Available at: https://encyclopedia.pub/entry/44613. Accessed February 28, 2026.
Airola, Carlo, Andrea Severino, Serena Porcari, William Fusco, Benjamin H. Mullish, Antonio Gasbarrini, Giovanni Cammarota, Francesca Romana Ponziani, Gianluca Ianiro. "Future Modulation of Gut Microbiota" Encyclopedia, https://encyclopedia.pub/entry/44613 (accessed February 28, 2026).
Airola, C., Severino, A., Porcari, S., Fusco, W., Mullish, B.H., Gasbarrini, A., Cammarota, G., Ponziani, F.R., & Ianiro, G. (2023, May 21). Future Modulation of Gut Microbiota. In Encyclopedia. https://encyclopedia.pub/entry/44613
Airola, Carlo, et al. "Future Modulation of Gut Microbiota." Encyclopedia. Web. 21 May, 2023.
Copy Citation
The human gut is inhabited by a multitude of bacteria, yeasts, and viruses. A dynamic balance among these microorganisms is associated with the well-being of the human being, and a large body of evidence supports a role of dysbiosis in the pathogenesis of several diseases. Given the importance of the gut microbiota in the preservation of human health, probiotics, prebiotics, synbiotics, and postbiotics have been classically used as strategies to modulate the gut microbiota and achieve beneficial effects for the host. Besides, several molecules not typically included in these categories have demonstrated a role in restoring the equilibrium among the components of the gut microbiota.
eubiotics
microbiota
dysbiosis
rifaximin
microbiota modulation
gastrointestinal diseases
1. Introduction
Interest in the human gut microbiota and its potential impact on human health has significantly increased since the development of metagenomic technologies that allow for the deep sequencing of microbial genomes. Trillions of bacteria live in the gut microbiota of the human digestive system and are established as a very complex environment. Beginning at birth, a wide range of genetic, nutritional, and environmental variables influence the composition of the gut microbiome [1]. After the beginning of the 20th century, Metchnikoff proposed that the human gut microbiome plays a part in both health and sickness [2]. Subsequently, the introduction of new molecular technologies that emerged in metagenomics spread a new perspective on microbiota research. The so-called “microbiota revolution” enabled the discovery of crucial links between pathogenic diseases and gut microbes [3].
The gut microbiota is composed of bacteria (generally including Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia phila), yeasts, and viruses [4][5][6]. The gut bacteria perform a variety of functions, including vitamin production, pathogen defense, immune response stimulation, metabolism regulation and drug absorption [6]. High taxonomic diversity, microbial gene richness, and a stable core microbiota are frequently observed in healthy microbiota communities [6]. However, the relative distribution of microorganisms varies between individuals, even within the same person. Nearly every element of the host can be impacted by the microbiota, and dysbiosis is linked to a wide range of illnesses. In particular, dysbiosis has been implicated in cardiovascular and respiratory diseases, inflammatory bowel diseases, liver disorders, a variety of neoplasms, and metabolic illnesses via the intensification of a chronic inflammatory state [7][8][9]. It has also been suggested that gut microbes play a part in preserving the homeostasis of the gut–brain axis [9]. It has been supposed that different molecular patterns enhance microbiota-associated pathogenic states. Recently, it was discovered that the microbiota produces small molecules called genotoxins. In particular, the family of indolimines generated by the M. morganii strains associated with IBD–colorectal cancer can enhance colon carcinogenesis in mice and increase intestinal permeability [10]. Other small-molecule metabolites of Gram-positive and Gram-negative bacteria, such as those from Clostridium perfringens and Clostridium ramosum strains, directly damage DNA and cause the expression of double-strand break markers—as well as cell-cycle arrest—in epithelial cells [10]. Alterations in intestinal microbiota can therefore be correlated to pathological states, both through a non-specific mechanism of chronic inflammation and through specific molecular patterns (Figure 1); therefore, it is essential to achieve the ability to both modulate complex microbial communities and target individual members within these communities.
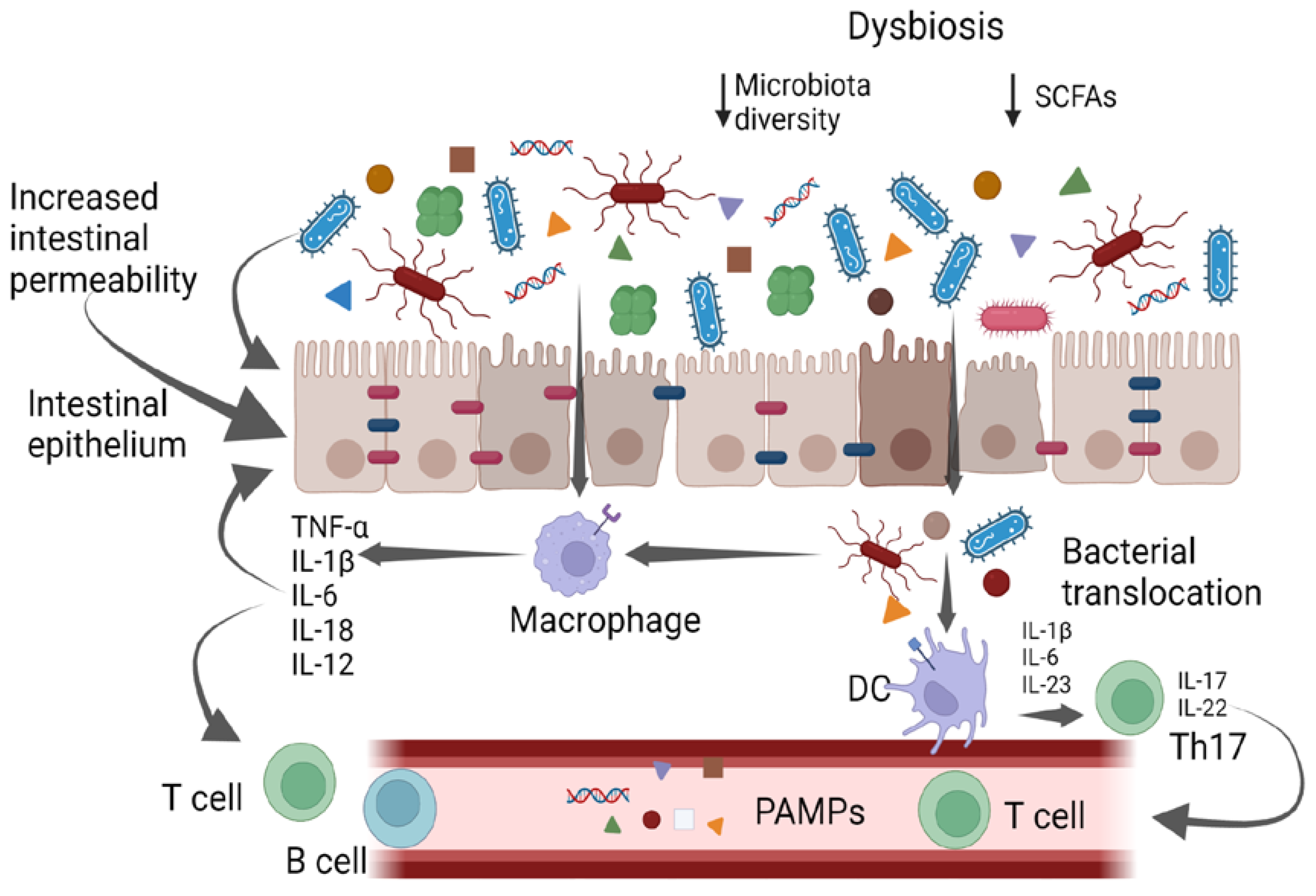
Figure 1. Gut bacteria and systemic inflammation. PAMPs—pathogen-associated molecular patterns; DCs—dendritic cells.
The intestinal mucosa provides a selective, permeable barrier for nutrient absorption and protection from external factors. Pathogens, xenobiotics, and food can disrupt the intestinal barrier, while genetic and immune factors predispose individuals to gut barrier dysfunction. The gut microbiota participates in regulating the integrity and function of the intestinal barrier in a homeostatic balance in various ways: opposing colonization by pathogens, promoting the differentiation of regulatory T (Treg) cells (which induce tolerance to lumen antigens), and stimulating B cells to secrete immunoglobulin (Ig)A to avoid bacterial translocation. Commensals also convert dietary fiber into short-chain fatty acids (SCFAs), which protect the gut barrier in various ways, including by providing energy for colonocytes and stimulating the production of mucus, antimicrobial proteins, and Treg cells. Dysbiosis and chronic disruption of the gut barrier can lead to the translocation of microbial components, the activation of pro-inflammatory patterns, and the production of systemic, low-grade inflammation [11][12][13].
To date, a multitude of molecules with a modulatory effect on gut microbiota have been developed [14][15][16][17][18]. Most of these agents are prebiotics, probiotics, synbiotics, or postbiotics [14][15][16][17][18]. However, some molecules known for their antimicrobial effect have been shown to improve the balance of the gut microbiota [19]. Due to their properties, these agents could not be included in the previously mentioned therapeutic groups [19]. Nonetheless, rather than being antimicrobial, their activity may be classified as eubiotic [19].
2. Eubiotics: Drugs to Modulate the Gut Microbiota
Prebiotics and probiotics have historically played a crucial role in the regulation of the gut microbiota. Prebiotics are typically non-digestible food components that selectively encourage the growth and activity of a small number of bacteria in the digestive tract [14]. Probiotics are live microorganisms which, when administered in adequate amounts, confer a health benefit to the host [15]. Synbiotics (a combination of prebiotics and probiotics) and postbiotics (inanimate microorganisms and/or their components that confer a health benefit to the host) have been recently introduced, showing a beneficial effect upon the dysbiotic state [16][17][18].
Nevertheless, pharmacological agents not included in the above-mentioned groups demonstrate an interesting ability to alter microbiota, preventing dysbiosis, with a significant effect on associated diseases. Antimicrobials, natural compounds, and even metabolites of the same gut microbiota have been considered. A new group, eubiotics, has been proposed [20][21]. All these molecules have a somewhat antimicrobial activity; however, rather than determining a global depletion in bacteria abundance, the antimicrobial activity leads to a modulation of the composition of microbiota, which can be beneficial for the host [20][21].
2.1. Rifaximin: New Perspectives for an Old Antibiotic
During the last half of the 20th century, a number of new antibacterial agents came into clinical use, providing clinicians with a variety of options when treating many types of infectious diseases; among these options, antibiotics played a major role [22]. In addition to their beneficial effect against pathogenic bacteria, antibiotics are associated with significant short- and long-term alterations in the composition of the human microbiota [23]. The way an antibiotic affects the gut microbiota depends largely on its class, as well as the composition of the microbiota in the gut before the antibiotic is administered [24].
The use of an antibiotic results in a decrease in alpha (α) diversity, which measures the variety of bacterial taxa present in a given individual’s microbiota, and in beta (β)-diversity, which is a marker of the homogeneity of bacterial relative abundance [24][25]. Since suppressing susceptible microbes can create an ecological niche for opportunistic pathogenic bacteria that are resistant to the antibiotic, increasing the host’s susceptibility to post-antibiotic infection, the majority of studies focus on the dysbiotic effect of antibiotics [23][26][27]. Additionally, it has been demonstrated that antibiotics enrich phage-encoded genes, which transfer resistance to the administered drug and unrelated antibiotics and encourage interactions between phages and bacteria, thereby enhancing the exchange of resistance genes [28].
Nonetheless, some antibiotics have shown an intriguing function in the clinical modulation of the gut microbiota. Taking into account changes in bacterial abundance, the non-systemic antibiotic rifaximin, which has bactericidal and bacteriostatic activity against both aerobic and anaerobic bacterial species, has demonstrated promising results [29]. Rifaximin also has bile-acid-dependent solubility, which increases its effectiveness in the small intestine while inhibiting colonic bacteria only moderately [30]. Rifaximin irreversibly binds the bacterial DNA-dependent RNA polymerase, inhibiting bacterial protein synthesis [31]. To date, rifaximin is usually administered with beneficial effects in the management of diseases associated with an alteration in the gut microbiota, such as irritable bowel syndrome (IBS), diverticular disease, and hepatic encephalopathy (HE) [32][33][34]. Orally administered rifaximin determines minimal changes in the composition of gut microbiota, promoting the growth of bacterial species with a beneficial impact [29]. In addition, rifaximin modulates the inflammatory response by upregulating the expression of NF-kB via the pregnane X receptor [35][36] and downregulating the pro-inflammatory cytokines interleukin-1B and tumor necrosis factor alpha (TNFα) [37][38]. As observed in a clinical trial based on a multi-tagged pyrosequencing analysis, rifaximin modulates the networks among several bacteria (Enterobacteriaceae, Bacteroidaceae, Veillonellaceae, Porphyromonadaceae, and Rikenellaceae) and bacterial metabolites when administered to patients with mild HE [39]. An analysis of the serum of treated patients revealed an increase in saturated and unsaturated fatty acids [39]. Furthermore, rifaximin increased bacterial diversity, the Bacteroidetes/Firmicutes ratio, and the abundance of Faecalibacterium prausnitzii, a butyrate producer with potent anti-inflammatory properties, in a second smaller study involving 15 patients with IBS [40]. An analysis of gut microbiota diversity showed that the clinical response was associated with a slight increase in α-diversity [40]. Treatment with 1200 mg of rifaximin daily for 10 days increased the abundance of Lactobacilli in another report on 19 patients with various gastrointestinal and liver disorders (inflammatory bowel disorder, IBS, diverticular disease, and HE), with no effects upon the α-diversity of the gut microbiota [20]. A recent clinical trial on patients with HE demonstrated that daily treatment with 1200 mg of rifaximin increased the abundance of Proteobacteria while decreasing the abundance of Firmicutes [41]. A metagenomic analysis highlighted that a decrease in the prevalence of Veillonella, Haemophilus, Streptococcus, Parabacteroides, Megamonas, Roseburia, Alistipes, Ruminococcus, and Lactobacillus was also associated with rifaximin administration. Despite this trend of bacterial abundance modification, the stability of the gut microbiota was not affected by rifaximin administration, and its minimal antibacterial effect in the intestine was confirmed in this report [41]. While the overall composition of the gut microbiota is unaffected by the treatment, rifaximin causes a relative rather than an absolute change in the abundance of these helpful bacteria. This might be because some microbes that are sensitive to rifaximin experienced a minimal reduction in abundance that is not statistically significant while others, such as Lactobacilli, developed resistance [20]. Interestingly, Brigidi et al. showed a significant initial decrease in the fecal abundance of Lactobacilli in individuals with mild or severe ulcerative colitis who were receiving a higher dosage of rifaximin (i.e., 1800 mg daily). However, the abundance of Lactobacilli returned after three cycles of therapy that each lasted 10 days. Moreover, Lactobacilli, which were particularly susceptible to rifaximin at the beginning of the study, developed resistance to the drug (to a mean value of 12 μg/mL) [42].
In order to improve the pharmacological approach and incorporate it into therapeutic options, it is necessary to investigate the mechanisms that underlie the clinical impact exerted by this beneficial gut microbiota perturbation.
2.2. The Multi-Layered Mechanisms of Rifaximin
Rifaximin, as previously mentioned, differs from other antibiotics as it does not affect the gut microbiota’s overall composition. Indeed, rifaximin increases the abundance of beneficial gut bacteria and promotes metabolic modifications, balancing the relationship between the host and the bacteria with a well-recognized positive clinical effect.
Although the underlying biological mechanisms are not fully understood, some metabolic networks may be suggested. Rifaximin treatment protected mice exposed to malathion from stress-induced oxidative damage in a pre-clinical study [43]. In the gut microbiota of rifaximin-treated mice, acetate- and propionate-producing bacteria, such as Lactobacillus spp., Bacteroides spp., Prevotella spp., Streptococcus spp., Phascolarctobacterium succinatutens, and Negativicutes spp., were found in high concentrations [43]. Therefore, treatment with rifaximin effectively increased the production of short-chain fatty acids (SCFAs) by modulating the gut microbiota [44]. Dupraz et al. showed that intestinal gamma-delta (γδ) T cells in mice and humans produce less IL-17 and IL-22 when microbiota-produced SCFAs, particularly propionate, are present. This results in a decrease in the inflammatory state [45]. Furthermore, the administration of rifaximin was linked to an increase in the expression of peroxisome proliferator-activated receptor gamma coactivator-1 alpha, a key metabolic regulator whose expression is elicited by SCFAs [43][46]. As a result, SCFAs might be the means through which rifaximin reduces systemic inflammation and exerts its beneficial effect. Another preclinical model highlighted rifaximin’s function in controlling systemic inflammatory responses. By inhibiting the activation of the TLR-4/NF-B signaling pathway and downregulating inflammatory factors such as TNF-α, IL-6, IL-17A, and IL-23, rifaximin significantly diminished the severity of clinical disease in mice with proteoglycan-induced ankylosing spondylitis. In this instance, a microbiological analysis showed an increase in Lactobacillus and Bacteroides, with a higher Bacteroidetes/Firmicutes ratio [47]. Additionally, in vitro studies performed in human gut epithelial cells indicated that rifaximin decreases apoptosis and increases tight junction protein expression by activating the TLR4, MyD88, and NF-kB pathways [48]. Meanwhile, increased populations of bacteria such as Lactobacilli have been shown to reduce the production of pro-inflammatory cytokines and TNF-α to prevent the spread of pathogenic bacteria and to modulate intestinal permeability [49][50][51].
As a result, rifaximin not only creates an anti-inflammatory environment by regulating intestinal metabolic pathways but also helps to reestablish the function of the intestinal barrier by reducing intestinal permeability, preventing the translocation and overgrowth of bacteria that could be harmful to the individual [21].
Patel et al. proposed that these preclinical implications could be transferred to a human model in a recent randomized controlled clinical trial [52]. In comparison to the placebo group, 90 days of rifaximin treatment decreased the plasma levels of tumor necrosis factor alpha (TNF-α) and circulating neutrophil TLR-4 expression in patients with cirrhosis and hepatic encephalopathy [52]. Additionally, metagenomic quantification techniques showed that rifaximin reduced the levels of sialidase-rich species that degrade mucin (i.e., Streptococcus spp., Veillonella atypica and parvula, Akkermansia, and Hungatella), preventing the so-called “oralization” of the gut microbiota [52]. These bacteria are present in the oral microbiota, but they are also more prevalent in the gut microbiota in conditions such as cirrhosis [53]. Sialidase alters the intestinal permeability by degrading O-glycans in the gut mucin barrier. Rifaximin also promoted an intestinal microenvironment rich in TNF-α and interleukin-17E (IL-17) [52]. It has been demonstrated that IL-17E (also known as IL-25), unlike other isoforms of IL-17, is implicated in the integrity of the gut barrier [54][55]. Intestinal tuft cells produce IL-17E in response to indole-acetic propionic acid, a tryptophan metabolite involved in gut homeostasis and anti-inflammatory pathways mediated by IL-10 [54][55]. It is also intriguing to note that plasma TNF-α levels decreased while intestinal TNF-α increased, demonstrating how the maintenance of gut homeostasis and the control of systemic inflammation can be achieved by increasing an inflammatory cytokine in a specific microenvironment. Clinically, rifaximin treatment resulted in a recovery of neurocognitive function, in addition to the complete resolution of hepatic encephalopathy. Additionally, rifaximin allowed for the preservation of beta (β) diversity in the gut microbiota, whereas the placebo group saw a decrease in this metric [52].
Rifaximin demonstrates great potential for maintaining the balance between the host and the gut microbiota. Molecular and microbiological data are strongly consistent with clinical evidence. Rifaximin’s effectiveness, however, may be impacted by various disruptions to the gut homeostasis that are related to various pathological conditions. On the other hand, rifaximin is a versatile drug because it has the ability to restore balance instead of acting on a single target. Regarding one of the main issues with antibiotic therapy, rifaximin showed no long-term effects, and its impact on bacterial resistance appears to be absolutely negligible [41]. The eubiotic effects of rifaximin are summarized in Figure 2.
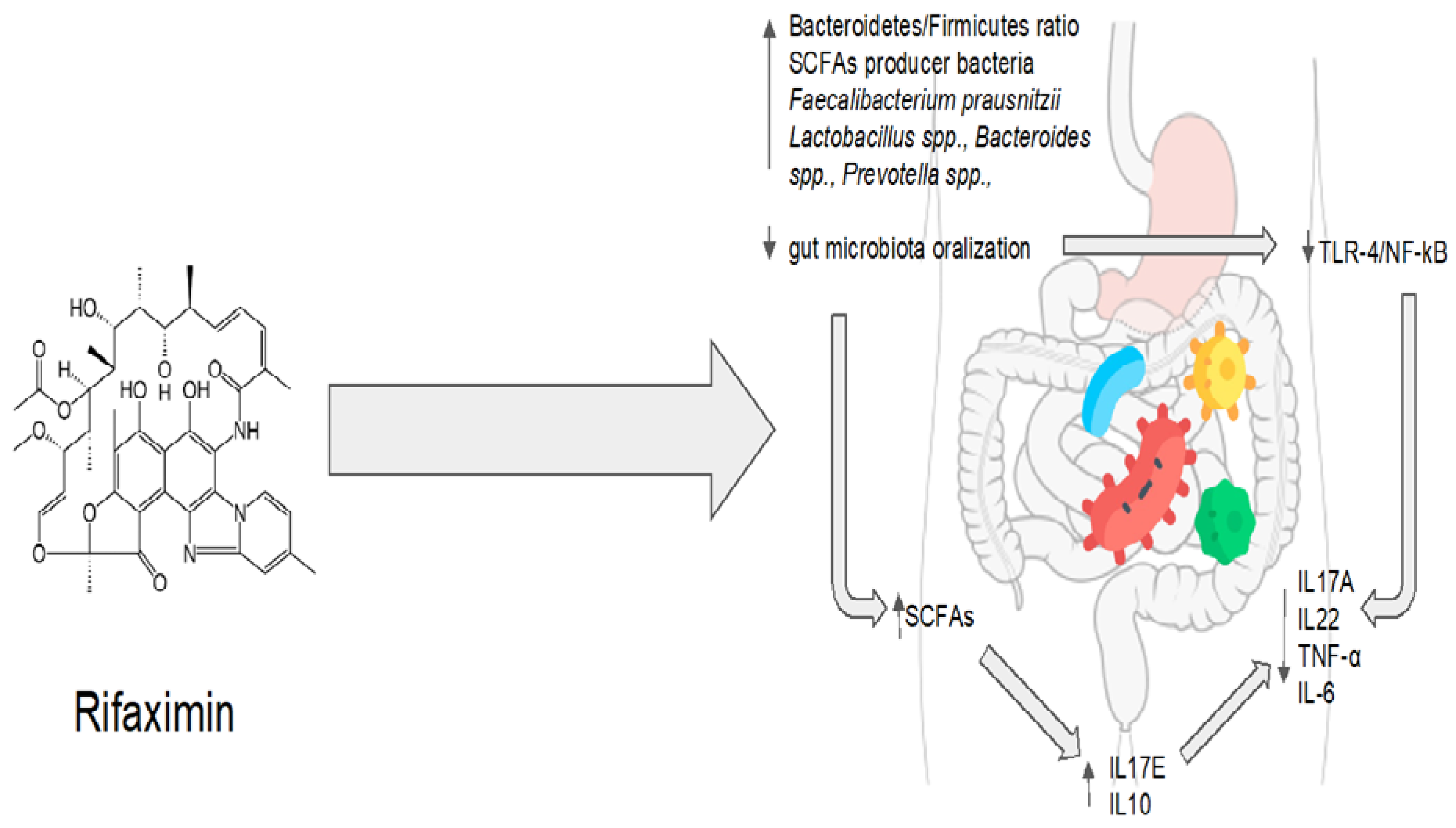
Figure 2. Rifaximin’s eubiotic effects. IL17A: interleukin 17A; IL22: interleukin 22; TNF-α: tumor necrosis factor alpha; IL-6: interleukin 6; IL10: interleukin 10; IL17E: interleukin 17E.
2.3. Other Antimicrobial Agents: Triclosan
Triclosan is a nonionic broad-spectrum antimicrobial agent. Its chemical name is 2,4,4′-trichloro-2′-hydroxydiphenyl ether. It is widely used in toothpaste, food storage containers, medical products, personal care products, and plastic cutting boards [56]. Triclosan acts as a detergent, directly disrupting the integrity of the bacterial membrane [57]. Moreover, it interferes with the synthesis of bacterial fatty acids by inhibiting the enoyl-acyl carrier protein (enoyl-ACP) reductase [58]. At low concentrations, it limits bacterial growth, while at high concentrations, it is bactericidal [59].
Since its introduction, numerous studies have examined the effects of triclosan on living organisms to determine its safety. A randomized study of triclosan-containing household and personal care products carried out to evaluate the safety of triclosan showed that triclosan exposure did not induce global reconstruction or the loss of microbial diversity in the gut microbiota [60]. Nevertheless, because triclosan-containing products included toothpaste, the oral microbiota was also examined. So far, even in sites where triclosan was applied directly, no significant differences in α-diversity were detected [60]. Triclosan seems to not have an antibacterial effect at the dosage used in household and personal care products, despite the fact that this effect may be present at higher concentrations and for longer periods of exposure [60][61].
Another preclinical model sought to define perturbations to the gut microbiota caused by triclosan. Microbiota composition was evaluated at three, twenty-one, and fifty-two weeks after the administration of a low dose of triclosan. Following exposure to triclosan (50 mg/kg/day), the abundance of Bacteroidetes increased at week 52. At the same dose after 52 weeks, triclosan slightly (but not significantly) reduced the abundance of Firmicutes. At 21 and 52 weeks of age, triclosan decreased the levels of Akkermansia muciniphila at the species level. Low doses of triclosan increased α-diversity after three weeks when compared to the control group [62]. Triclosan resistance mechanisms include changes in the enoyl-ACP reductase and efflux pumps. However, despite the widespread use of products containing triclosan, the community’s overall resistance and cross-resistance rates are low [63].
Triclosan consequently gained attention as a modulator of the gut microbiota, and it was considered as a drug for the treatment of dysbiosis in a recent preclinical study [64]. Mice received a continuous high-fat diet for 20 weeks and were then treated with triclosan at a dose of 400 mg/kg/d for the final 8 weeks [64]. In mice fed a high-fat diet, triclosan increased the ratio of Bacteroidetes/Firmicutes and decreased the number of pathogenic Gram-negative bacteria, such as Helicobacter, Erysipelatoclostridium, and Citrobacter, as shown by a metagenomic analysis [64]. Triclosan also increased the relative abundance of Lactobacillus, Bifidobacterium, and Lachnospiraceae, which protect against abnormal metabolic processes [64]. The increase in α- and β-diversity also demonstrated triclosan’s improvement of bacterial diversity and richness [64].
On the other hand, a cohort study showed that triclosan exposure causes a decrease in the diversity of the gut microbiota in breast-fed infants [65]. This study, however, is not representative of the general population due to the small sample size and the high vulnerability of the infant gut microbiota to antimicrobial agents. Additionally, rather than thinking of triclosan as a perturbator of a balanced microenvironment, it would be interesting to consider it as a modulator of the gut microbiota in patients with a state of gut dysbiosis [65]. Indeed, triclosan is employed in periodontal disease, which could be considered a model of oral dysbiosis [66]. Nonetheless, oral dysbiosis, which is characterized by an increased abundance of Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans, has been associated with the impairment of the gut microbiota equilibrium and the loss of gut microbial diversity [67][68]. Moreover, these oral pathobionts have been associated with NAFLD and NASH progression in preclinical models [67][69]. As a consequence, periodontal therapy can improve oral and gut dysbiosis in patients with chronic liver diseases [70]. In particular, periodontal therapy in cirrhotic individuals reduces blood levels of inflammatory cytokines and lipopolysaccharide (LPS), with beneficial effects upon both quality of life and in mitigating the development of hepatic encephalopathy [70]. Triclosan, as a treatment for periodontal disease [71], has been shown to reduce the production of pro-inflammatory cytokines (such as IL-8, IL-1 α, and TNFα) in a human epithelial cells, monocytes, and fibroblast cultures exposed to LPS [72]. More specifically, triclosan treatment inhibits the TLR-4 pathway by inducing the microRNA miR146a to downregulate IRAK1 and TRAF6 proteins. Conversely, triclosan exposure increases the epithelial cells’ production of other bioactive anti-microbial molecules, such as β-dsefensins [72]. Interestingly, β-defensins have been proposed as a key factor in the physiological homeostasis between the host and the microbiome [73].
In conclusion, triclosan, an antimicrobial agent with a long history, has been suggested to have a positive impact on the microbiota in the human gut. In addition to its well-known antibacterial action, triclosan could have a pleiotropic effect on different cell types, directly orchestrating physiological homeostasis between microbiota and their host. However, more investigations are necessary.
Triclosan’s eubiotic effects are summarized in Figure 3.
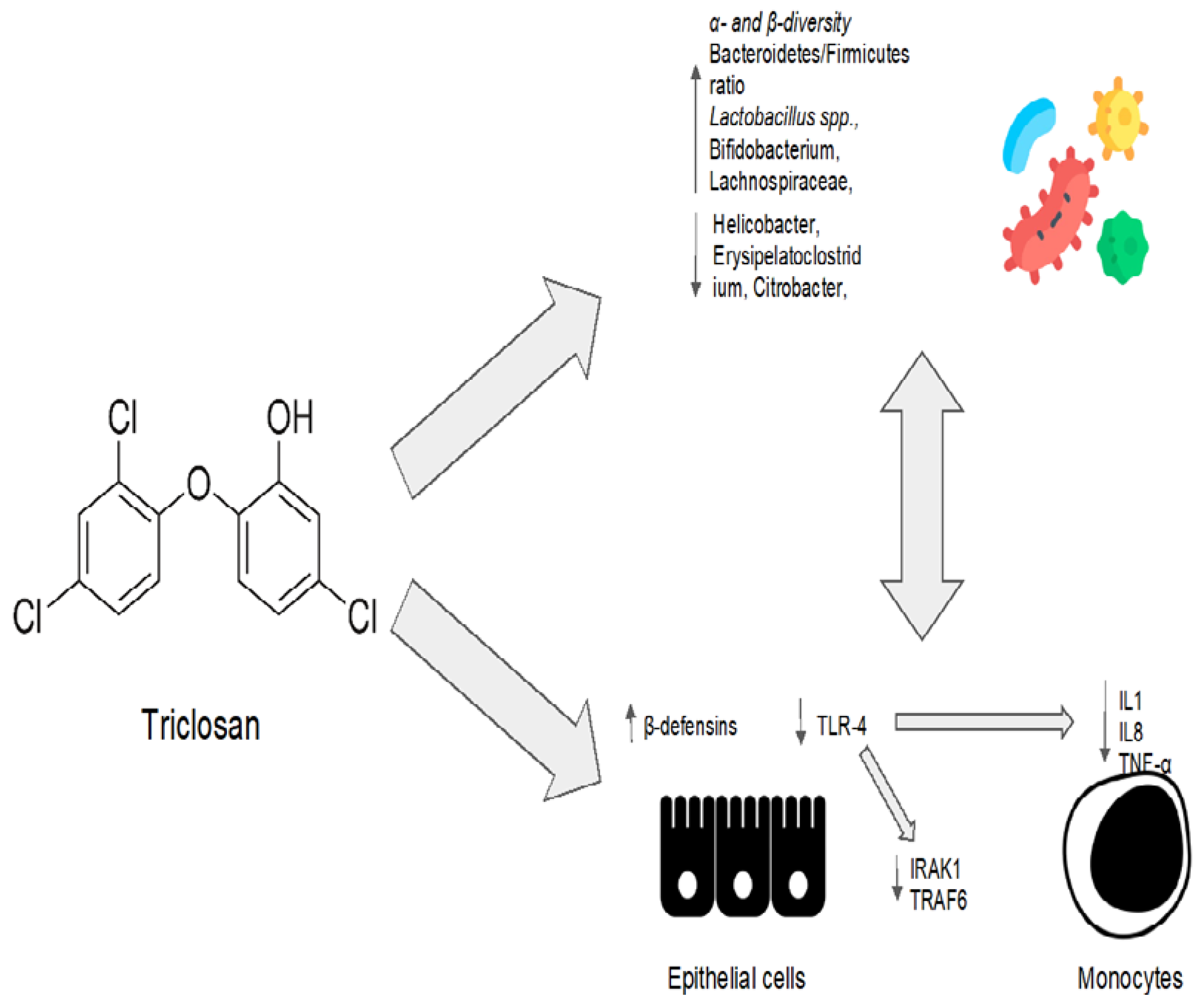
Figure 3. Triclosan’s eubiotic effects. IL1: interleukin 1; IL8: interleukin 8; TNF-α: tumor necrosis factor alpha; IRAK1: interleukin 1 receptor-associated kinase 1; TRAF6: TNF-receptor-associated factor 6.
2.4. Natural Products: Promising Agents for the Modulation of Microbiota
Evodiamine is an alkaloid, mainly present in the Evodia Fructus, which is officially listed in the Chinese Pharmacopoeia as a remedy for patients suffering from viral hepatitis, cholangitis, and gastric ulcers, among other disorders [74]. Evodiamine has been demonstrated to have an antimicrobial activity in vitro, inhibiting bacterial topoisomerase I [74].
Evodiamine’s impact on intestinal inflammation and the gut microbiota was examined in a preclinical study [75]. In order to create a intestinal inflammatory tumor mouse model, azomethane/sodium dextran sulfate was used [75]. The tumor model was then treated with evodiamine and 5-aminosalicylic acid. Evodiamine and 5-aminosalicylic acid both prevented the growth of tumors and induced the death of tumor cells [75]. A quantitative polymerase chain reaction analysis showed that evodiamine and 5-aminosalicylic acid reduced the number of Enterococcus faecalis and Escherichia coli while increasing the abundance of Bifidobacterium, Campylobacter, and Lactobacillus when compared to the control group [75]. The IL6/STAT3/P65 signaling pathway was inhibited, and levels of inflammatory factor, d-lactic acid, and serum endotoxin were all significantly decreased in the evodiamine group [75]. Evodiamine also showed efficacy in preventing colorectal tumors in a mouse model of chemically induced colitis [76]. In this model, evodiamine increased the abundance of SCFA-producing bacteria, inhibiting the harmful bacteria [76]. After evodiamine treatment, histological analysis showed a remarkable reversion of intestinal epithelial structure destruction, inflammatory cells infiltration, and crypt loss. The intestinal barrier was also restored, with an increased expression of occludin, zonula occludens-1, and E-cadherin [76][77]. Additionally, evodiamine decreased the expression of pro-inflammatory genes involved in the Wnt signaling pathway, the Hippo signaling pathway, and the IL-17 signaling pathway [76]. This anti-inflammatory effect could be mediated by the downregulation of the nuclear factor-kappa B (NF-κB) signal and the inhibition of NLRP3 inflammasome activation, as shown in another sodium-dextran-sulfate-induced colitis murine model [77].
The effect of evodiamine was also evaluated on H. pylori in an in vitro gastric adenocarcinoma model [78]. Evodiamine treatment decreased the bacterial production of the type IV secretion system components and the system subunit protein A protein and the expression of cytotoxin-associated antigen A (CagA) and vacuolating cytotoxin A (VacA). This resulted in a reduction in CagA end VacA proteins in tumor cells. In addition, evodiamine specifically prevented the H. pylori-infection-induced stimulation of signaling proteins such as NF-κB and the mitogen-activated protein kinase (MAPK) pathway. As a result, IL-8 secretion in tumoral cells was reduced [78].
Evodiamine and berberine were combined in order to determine their effect on the gut microbiota of rats fed a high-fat diet [79]. The treatment with evodiamine and berberine increased the diversity of the gut microbiota globally [79]. The gut microbiota profiles were determined via the high-throughput sequencing of the bacterial 16S ribosomal RNA gene [79]. In comparison to the control group, there were higher proportions of SCFA-producing bacteria (Lactobacillus, Prevotella, Ruminococcaceae, and Bacteroides). In contrast, the key bacteria responsible for the imbalance in the gut microbiota in the group with a high-fat diet were Fusobacteria and Lachnospiraceae [79]. The high-fat-diet group’s Firmicutes/Bacteroidetes ratio was significantly higher than that of the control group [79]. The administration of evodiamine and berberine reverted the increasing trend of the ratio. Evodiamine–berberine also lowered the intestinal submucosal edema typical of a high-fat diet and the mucosal inflammatory cell infiltration [79]. As a result, the rats treated with evodiamine and berberine showed a significant reduction in body weight, as well as plasma triglyceride and total cholesterol levels [79]. Liver injury (measured via plasma levels of aspartate aminotransferase, alanine aminotransferase, and gamma-glutamyl transpeptidase) was significantly reduced [79]. Evodiamine’s eubiotic effects are summarized in Figure 4.
The capacity of other substances to alter the microbiota has been researched in relation to their potential role in the treatment of colorectal cancer. Natural products, and in particular natural extracts, are frequently the source of anticancer agents. In mice with heterotopic xenograft colorectal cancer, an ethanol extract of Euphorbia lathyris was found to reduce tumor size [80]. When compared to mice without colorectal cancer, a gut microbiota analysis showed a significant decrease in the abundance of Lactobacilli, with predominance of colitogenic bacteria such as Akkermansia and Turicibacter. Turicibacter disappeared after treatment with an ethanol extract of Euphorbia lathyris, and Lactobacillus abundance recovered [80]. Nevertheless, it is still unclear whether the Euphorbia lathyris extract impacted the microbiota directly or whether it modulated gut microorganisms as a result of its anti-neoplastic effects.
Propolis is a resinous material collected by bees from the buds and resins of plants and mixed with bee enzymes, pollen, and wax [81]. It has been used as a traditional remedy worldwide. In some countries, it is considered a complementary medicine, while it is regulated as a food supplement in others [82]. It contains a variety of molecular compounds, such as flavonoids, terpenes, phenolic acids, β-steroids, and the derivatives of sesquiterpenes, naphthalene, and stilbenes [83]. Its antioxidant, anti-inflammatory, and antimicrobial activities are probably due to its polyphenol contents. Concerning the microbiota, propolis has been demonstrated to modulate intestinal bacteria, resulting in an improvement in intestinal barrier function in diabetic rats [83]. However, due to the content variability of propolis, studies require the standardization of its content. A recent preclinical trial showed that a standardized polyphenol propolis mixture modulated the gut microbiota in an in vitro human model, determining an increase in the production of SCFAs [82]. A propolis mix with 7.21 g of total polyphenols/g was specifically used. The propolis extract’s polyphenols included quercetin, apigenin, pinobanskin, chrysin, pinocembrin, and galangin at defined dosages. The standardized polyphenol combination underwent an in vitro digestion process which mimicked human digestion before being fermented by fecal microbiota isolated from healthy adults and children, obese children, celiac children, and children with a food allergy. The production of SCFAs significantly increased following these processes, as shown by a combined chromatographic and UV detection technique. Particularly, samples from the allergic, obese, and celiac patients had significantly higher levels of acetic acid production than the samples from healthy people. On the other hand, compared to other samples, the samples from individuals with allergies had higher levels of propionic acid [82].
Polyphenols are primarily produced by plants as defense against pathogenic microorganisms [84]. As different microbes have different metabolisms, polyphenols appear to selectively inhibit bacterial growth. Tannic acid, for instance, did not affect the growth of Bifidobacterium infantis and Lactobacillus acidophilus, but it inhibited the growth of Clostridium clostridiiforme, E. coli ATCC 25,922, and Enterobacter cloacae ATCC 13,047 in anaerobic conditions. Tannic acid is a potent iron chelator, and its ability to inhibit bacterial growth may be due to the fact that it deprives bacteria of the iron that they require for metabolism (Bifidobacterium infantis and Lactobacillus acidophilus do not) [85]. Other polyphenols have a more specific mechanism of action. Epigallocatechin directly binds the peptidoglycan in the S. aureus cellular wall, causing osmotic damage and bacterial death [86]. Furthermore, tea polyphenols exert direct damage on the outer and inner membranes of S. marcescens, dramatically increasing cellular permeability and thus inhibiting bacterial growth [87].
One of the most investigated polyphenols is resveratrol, a phytoalexin present in many plants such as grapes, peanuts, and berries [88]. Resveratrol has demonstrated antimicrobial and antifungal activity by preventing the formation of biofilms [89][90][91]. Resveratrol, previously administered to mice receiving intrarectal treatment with the oxidizing agent 2,4,6-trinitrobenzenesulfonic acid, demonstrates a beneficial effect on the gut microbiota. Genomic DNA analysis and 16S rRNA gene sequencing revealed that oxidative stress increased species such as Bacteroides acidifaciens and decreased species such as Ruminococcus gnavus and Akkermansia muciniphila, according to an analysis of stool microbiota. The administration of resveratrol returned the gut bacteria to their homeostatic levels and increased the production of butyric acid. A mesenteric lymph node analysis via cytometry showed a significant increase in the percentages of both anti-inflammatory CD4+FOXP3+ Tregs and CD4+IL10+ in the resveratrol-treated group. Meanwhile, inflammatory Th1/Th17 cells were suppressed, and mucosal inflammation was also significantly reduced [92]. A microarray analysis of microRNA profiles in mesenteric lymph node cells highlighted that resveratrol treatment was associated with miR-31, Let-7a, and miR-132 downregulation [93]. All these microRNAs target anti-inflammatory T cells. In particular, miR-31 inhibits the production of FoxP3 [94]. It is noteworthy to observe that miR-31 expression in tissues connected to the disease is considerably greater among individuals with ulcerative colitis [93]. Moreover, by lowering inflammation and regulating hepatic lipid metabolism, resveratrol has been demonstrated to lower the risk of non-alcoholic fatty liver disease (NAFLD). Resveratrol treatment reduced hepatic steatosis in male mice fed with a high-fat diet and modified the composition of the gut bacteria by enhancing the growth of SCFA-producing bacteria such as Allobaculum, Bacteroides, and Blautia [95]. A clinical trial investigated the impact of resveratrol on the fecal microbiota of overweight men and women [96]. In comparison to women, men have a higher fecal abundance of Bacteroidetes. Resveratrol (282 and 80 mg/day, respectively) and another phenolic compound called epigallocatechin-3-gallate were combined for 12 weeks [96]. Men experienced a decline in the relative abundance of Bacteroidetes, but women did not. Administration had no effect on firmicutes, actinobacteria, gammaproteobacteria, Akkermansia muciniphila, sulfate-reducing bacteria, or acetogenic bacteria in either men or women. By using indirect calorimetry, it was demonstrated that fat oxidation was increased in men compared to women [96]. These findings highlight the importance of a gut microbiota modulator’s ability to restore an altered equilibrium. Instead of acting against a specific target, a good modulator suppresses a deregulated element until it resumes its homeostatic role [96]. Resveratrol’s eubiotic effects are summarized in Figure 4.
Other relevant substances have been discovered to be elevated in the serum of people eating a Mediterranean diet. Indole-3-propionic acid is one specific example. This tryptophan metabolite, which is produced by gut bacteria, induces tuft cells to produce IL-17E, which has a modulatory effect on the gut microbiota, preventing lipoperoxidation and oxidative stress injury and reducing the synthesis of proinflammatory cytokines [97]. The administration of indole-3-propionic acid resulted in a significantly different composition of the intestinal microbiota in a sepsis mouse model, with an enrichment of the Bifidobacteriaceae family and a depletion of the Enterobacteriaceae family. In comparison to the control group, it resulted in lower serum inflammatory mediator levels and a higher survival rate [98]. This emphasizes once more how a compound that affects the gut microbiota can modify a systemic condition. In a different preclinical study, indole-3-propionic acid prevented the development of nonalcoholic steatohepatitis in mice fed a high-fat diet [99]. An analysis of the fecal microbiota via DNA sequencing techniques revealed a decrease in the Firmicutes/Bacteroidetes ratio as well as a decrease in the overall population of the pathobiont Streptococcus [99]. Rats fed a high-fat diet showed a loss of the normal villus structure of the ileum epithelium at the histological level. Treatment with indole-3-propionic acid improved the expression of tight junction proteins and restored the height of the ileum villus [99]. In addition, endotoxin plasma levels were lower in mice treated with indole-3-propionic acid than in the control group. As a result, when indole-3-propionic acid was administered, a significant histological reduction in steatohepatitis was seen [99]. Nonetheless, indole-3-propionic acid has been found to reverse dysbiosis induced by total abdominal irradiation in a mouse model, which was characterized by a decrease in the relative abundance of Lactobacillus and an increase in Bacteroides acidifaciens and Ruminococcus gauvreauii [100].
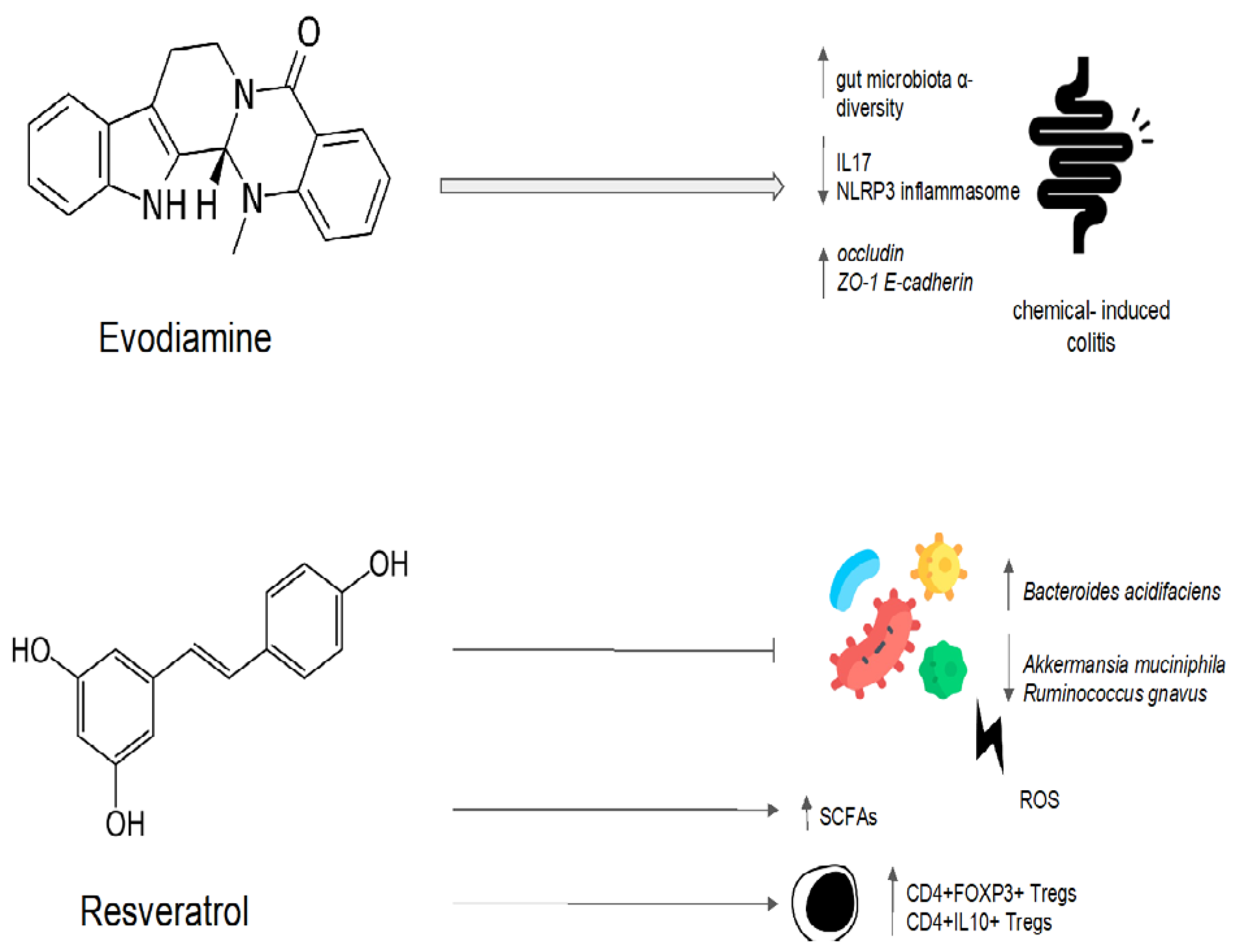
Figure 4. Possible mechanisms of the eubiotic effect of evodiamine and resveratrol. IL17: interleuchina-17; NLRP3: NOD-like receptor family, pyrin domain-containing protein 3; ROS: reactive oxygen species; ZO-1: zonula occludens-1; E-cadherin: epithelial cadherin.
References
- Gomaa, E.Z. Human gut microbiota/microbiome in health and diseases: A review. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2020, 113, 2019–2040.
- Metchnikoff, E. The Prolongation of Life: Optimistic Studies; Springer Publishing Company: New York, NY, USA, 2004.
- Blaser, M.J. The microbiome revolution. J. Clin. Investig. 2014, 124, 4162–4165.
- Ianiro, G.; Bruno, G.; Lopetuso, L.; Bartoli Beghella, F.; Laterza, L.; D’Aversa, F.; Gigante, G.; Cammarota, G.; Gasbarrini, A. Role of yeasts in healthy and impaired gut microbiota: The gut mycome. Curr. Pharm. Des. 2014, 20, 4565–4569.
- Ianiro, G.; Iorio, A.; Porcari, S.; Masucci, L.; Sanguinetti, M.; Perno, C.F.; Gasbarrini, A.; Putignani, L.; Cammarota, G. How the gut parasitome affects human health. Ther. Adv. Gastroenterol. 2022, 15, 17562848221091524.
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71.
- D’Aversa, F.; Tortora, A.; Ianiro, G.; Ponziani, F.R.; Annicchiarico, B.E.; Gasbarrini, A. Gut microbiota and metabolic syndrome. Intern. Emerg. Med. 2013, 8 (Suppl. 1), S11–S15.
- Bibbò, S.; Ianiro, G.; Dore, M.P.; Simonelli, C.; Newton, E.E.; Cammarota, G. Gut Microbiota as a Driver of Inflammation in Nonalcoholic Fatty Liver Disease. Mediat. Inflamm. 2018, 2018, 9321643.
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135.
- Cao, Y.; Oh, J.; Xue, M.; Huh, W.J.; Wang, J.; Gonzalez-Hernandez, J.A.; Rice, T.A.; Martin, A.L.; Song, D.; Crawford, J.M.; et al. Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science 2022, 378, eabm3233.
- Malesza, I.J.; Malesza, M.; Walkowiak, J.; Mussin, N.; Walkowiak, D.; Aringazina, R.; Bartkowiak-Wieczorek, J.; Mądry, E. High-Fat, Western-Style Diet, Systemic Inflammation, and Gut Microbiota: A Narrative Review. Cells 2021, 10, 3164.
- Martel, J.; Chang, S.-H.; Ko, Y.-F.; Hwang, T.-L.; Young, J.D.; Ojcius, D.M. Gut barrier disruption and chronic disease. Trends Endocrinol. Metab. 2022, 33, 247–265.
- Di Tommaso, N.; Gasbarrini, A.; Ponziani, F.R. Intestinal Barrier in Human Health and Disease. Int. J. Environ. Res. Public Health 2021, 18, 12836.
- da Silva, T.F.; Casarotti, S.N.; de Oliveira, G.L.V.; Penna, A.L.B. The impact of probiotics, prebiotics, and synbiotics on the biochemical, clinical, and immunological markers, as well as on the gut microbiota of obese hosts. Crit. Rev. Food Sci. Nutr. 2020, 61, 337–355.
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514.
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667.
- Vallianou, N.; Stratigou, T.; Christodoulatos, G.S.; Tsigalou, C.; Dalamaga, M. Probiotics, Prebiotics, Synbiotics, Postbiotics, and Obesity: Current Evidence, Controversies, and Perspectives. Curr. Obes. Rep. 2020, 9, 179–192.
- Mosca, A.; Abreu, A.T.A.Y.; Gwee, K.A.; Ianiro, G.; Tack, J.; Nguyen, T.V.H.; Hill, C. The clinical evidence for postbiotics as microbial therapeutics. Gut Microbes 2022, 14, 2117508.
- Rizzatti, G.; Ianiro, G.; Gasbarrini, A. Antibiotic and Modulation of Microbiota. J. Clin. Gastroenterol. 2018, 52 (Suppl. 1), S74–S77.
- Ponziani, F.R.; Scaldaferri, F.; Petito, V.; Paroni Sterbini, F.; Pecere, S.; Lopetuso, L.R.; Palladini, A.; Gerardi, V.; Masucci, L.; Pompili, M.; et al. The Role of Antibiotics in Gut Microbiota Modulation: The Eubiotic Effects of Rifaximin. Dig. Dis. 2016, 34, 269–278.
- Ponziani, F.R.; Zocco, M.A.; D’aversa, F.; Pompili, M.; Gasbarrini, A. Eubiotic properties of rifaximin: Disruption of the traditional concepts in gut microbiota modulation. World J. Gastroenterol. 2017, 23, 4491–4499.
- Powers, J. Antimicrobial drug development–The past, the present, and the future. Clin. Microbiol. Infect. 2004, 10, 23–31.
- Van Zyl, K.N.; Matukane, S.R.; Hamman, B.L.; Whitelaw, A.C.; Newton-Foot, M. Effect of antibiotics on the human microbiome: A systematic review. Int. J. Antimicrob. Agents 2021, 59, 106502.
- Gough, E.K. The impact of mass drug administration of antibiotics on the gut microbiota of target populations. Infect. Dis. Poverty 2022, 11, 76.
- Ianiro, G.; Tilg, H.; Gasbarrini, A. Antibiotics as deep modulators of gut microbiota: Between good and evil. Gut 2016, 65, 1906–1915.
- Ng, K.M.; Ferreyra, J.A.; Higginbottom, S.K.; Lynch, J.B.; Kashyap, P.C.; Gopinath, S.; Naidu, N.; Choudhury, B.; Weimer, B.C.; Monack, D.M.; et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013, 502, 96–99.
- Ferreyra, J.A.; Wu, K.J.; Hryckowian, A.J.; Bouley, D.M.; Weimer, B.C.; Sonnenburg, J.L. Gut microbiota-produced succinate promotes C. Difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 2014, 16, 770–777.
- Karami, N.; Hannoun, C.; Adlerberth, I.; Wold, A.E. Colonization dynamics of ampicillin-resistant Escherichia coli in the infantile colonic microbiota. J. Antimicrob. Chemother. 2008, 62, 703–708.
- Ponziani, F.R.; Gerardi, V.; Pecere, S.; D’aversa, F.; Lopetuso, L.; Zocco, M.A.; Pompili, M.; Gasbarrini, A. Effect of rifaximin on gut microbiota composition in advanced liver disease and its complications. World J. Gastroenterol. 2015, 21, 12322–12333.
- Darkoh, C.; Lichtenberger, L.M.; Ajami, N.; Dial, E.J.; Jiang, Z.-D.; DuPont, H.L. Bile acids improve the antimicrobial effect of rifaximin. Antimicrob. Agents Chemother. 2010, 54, 3618–3624.
- Hartmann, G.; Honikel, K.O.; Knüsel, F.; Nüesch, J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim. Biophys. Acta (BBA)-Nucleic Acids Protein Synth. 1967, 145, 843–844.
- Pimentel, M.; Lembo, A.; Chey, W.D.; Zakko, S.; Ringel, Y.; Yu, J.; Mareya, S.M.; Shaw, A.L.; Bortey, E.; Forbes, W.P. Rifaximin Therapy for Patients with Irritable Bowel Syndrome without Constipation ABSTRACT. N. Engl. J. Med. 2011, 364, 22–32.
- Bajaj, J.S.; Heuman, D.M.; Wade, J.B.; Gibson, D.P.; Saeian, K.; Wegelin, J.A.; Hafeezullah, M.; Bell, D.E.; Sterling, R.K.; Stravitz, R.T.; et al. Rifaximin improves driving simulator performance in a randomized trial of patients with minimal hepatic encephalo-pathy. Gastroenterology 2011, 140, 478–487.e1.
- Cuomo, R.; Barbara, G.; Annibale, B. Rifaximin and diverticular disease: Position paper of the Italian Society of Gastroenterology (SIGE). Dig. Liver Dis. 2017, 49, 595–603.
- Mencarelli, A.; Renga, B.; Palladino, G.; Claudio, D.; Ricci, P.; Distrutti, E.; Barbanti, M.; Baldelli, F.; Fiorucci, S. Inhibition of NF-κB by a PXR-dependent pathway mediates counter-regulatory activities of rifaximin on innate immunity in intestinal epithelial cells. Eur. J. Pharmacol. 2011, 668, 317–324.
- Mencarelli, A.; Migliorati, M.; Barbanti, M.; Cipriani, S.; Palladino, G.; Distrutti, E.; Renga, B.; Fiorucci, S. Pregnane-X-receptor mediates the anti-inflammatory activities of rifaximin on detoxification pathways in intestinal epithelial cells. Biochem. Pharmacol. 2010, 80, 1700–1707.
- Brown, E.L.; Xue, Q.; Jiang, Z.-D.; Xu, Y.; DuPont, H.L. Pretreatment of epithelial cells with rifaximin alters bacterial attachment and internalization profiles. Antimicrob. Agents Chemother. 2010, 54, 388–396.
- Amaya, E.; Caceres, M.; Fang, H.; Ramirez, A.T.; Palmgren, A.C.; Nord, C.E.; Weintraub, A. Extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a Neonatal Intensive Care Unit in León, Nicaragua. Int. J. Antimicrob. Agents 2009, 33, 386–387.
- Bajaj, J.S.; Heuman, D.M.; Sanyal, A.J.; Hylemon, P.B.; Sterling, R.K.; Stravitz, R.T.; Fuchs, M.; Ridlon, J.M.; Daita, K.; Monteith, P.; et al. Modulation of the Metabiome by Rifaximin in Patients with Cirrhosis and Minimal Hepatic Encephalopathy. PLoS ONE 2013, 8, e60042.
- Ponziani, F.; Scaldaferri, F.; de Siena, M.; Mangiola, F.; Matteo, M.; Pecere, S.; Petito, V.; Sterbini, F.P.; Lopetuso, L.; Masucci, L.; et al. Increased Faecalibacterium abundance is associated with clinical improvement in patients receiving rifaximin treatment. Benef. Microbes 2020, 11, 519–525.
- Yu, X.; Jin, Y.; Zhou, W.; Xiao, T.; Wu, Z.; Su, J.; Gao, H.; Shen, P.; Zheng, B.; Luo, Q.; et al. Rifaximin Modulates the Gut Microbiota to Prevent Hepatic Encephalopathy in Liver Cirrhosis Without Impacting the Resistome. Front. Cell. Infect. Microbiol. 2022, 11, 1427.
- Brigidi, P.; Swennen, E.; Rizzello, F.; Bozzolasco, M.; Matteuzzi, D. Effects of Rifaximin Administration on the Intestinal Microbiota in Patients with Ulcerative Colitis. J. Chemother. 2002, 14, 290–295.
- Omar, N.N.; Mosbah, R.A.; Sarawi, W.S.; Rashed, M.M.; Badr, A.M. Rifaximin Protects against Malathion-Induced Rat Testicular Toxicity: A Possible Clue on Modulating Gut Microbiome and Inhibition of Oxidative Stress by Mitophagy. Molecules 2022, 27, 4069.
- Li, H.; Xiang, Y.; Zhu, Z.; Wang, W.; Jiang, Z.; Zhao, M.; Cheng, S.; Pan, F.; Liu, D.; Ho, R.C.M.; et al. Rifaximin-mediated gut microbiota regulation modulates the function of microglia and protects against CUMS-induced depression-like behaviors in adolescent rat. J. Neuroinflammation 2021, 18, 254.
- Dupraz, L.; Magniez, A.; Rolhion, N.; Richard, M.L.; Da Costa, G.; Touch, S.; Mayeur, C.; Planchais, J.; Agus, A.; Danne, C.; et al. Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal γδ T cells. Cell Rep. 2021, 36, 109332.
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517.
- Yang, L.; Liu, B.; Zheng, J.; Huang, J.; Zhao, Q.; Liu, J.; Su, Z.; Wang, M.; Cui, Z.; Wang, T.; et al. Rifaximin alters intestinal microbiota and prevents progression of ankylosing spondylitis in mice. Front. Cell. Infect. Microbiol. 2019, 9, 44.
- Sánchez-Rabaneda, F.; Jáuregui, O.; Casals, I.; Andrés-Lacueva, C.; Izquierdo-Pulido, M.; Lamuela-Raventós, R.M. Liquid chromatographic/electrospray ionization tandem mass spectrometric study of the phenolic composition of cocoa (Theobroma cacao). J. Mass Spectrom. 2003, 38, 35–42.
- Zareie, M.; Johnson-Henry, K.; Jury, J.; Yang, P.-C.; Ngan, B.-Y.; McKay, D.M.; Soderholm, J.D.; Perdue, M.H.; Sherman, P.M. Probiotics prevent bacterial translocation and improve intestinal barrier function in rats following chronic psychological stress. Gut 2006, 55, 1553–1560.
- Thomas, C.M.; Hong, T.; Van Pijkeren, J.P.; Hemarajata, P.; Trinh, D.V.; Hu, W.; Britton, R.A.; Kalkum, M.; Versalovic, J. Histamine Derived from Probiotic Lactobacillus reuteri Suppresses TNF via Modulation of PKA and ERK Signaling. PLoS ONE 2012, 7, e31951.
- Llopis, M.; Antolin, M.; Carol, M.; Borruel, N.; Casellas, F.; Martinez, C.; Espín-Basany, E.; Guarner, F.; Malagelada, J.R. Lactobacillus casei downregulates commensals’ inflammatory signals in Crohn’s disease mucosa. Inflamm. Bowel Dis. 2009, 15, 275–283.
- Patel, V.C.; Lee, S.; McPhail, M.J.; Da Silva, K.; Guilly, S.; Zamalloa, A.; Witherden, E.; Støy, S.; Vijay, G.K.M.; Pons, N.; et al. Rifaximin-α reduces gut-derived inflammation and mucin degradation in cirrhosis and encephalopathy: RIFSYS randomised controlled trial. J. Hepatol. 2021, 76, 332–342.
- Woodhouse, C.; Singanayagam, A.; Patel, V.C. Modulating the gut–liver axis and the pivotal role of the faecal microbiome in cirrhosis. Clin. Med. J. R. Coll. Physicians Lond. 2020, 20, 493–500.
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; et al. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. Am. J. Pathol. 2018, 188, 1183–1194.
- Chen, L.; Yang, Y.; Sun, S.; Xie, Y.; Pan, C.; Li, M.; Li, C.; Liu, Y.; Xu, Z.; Liu, W.; et al. Indolepropionic acid reduces obesity-induced metabolic dysfunction through colonic barrier restoration mediated via tuft cell-derived IL-25. FEBS J. 2022, 289, 5985–6004.
- Sinicropi, M.S.; Iacopetta, D.; Ceramella, J.; Catalano, A.; Mariconda, A.; Pellegrino, M.; Saturnino, C.; Longo, P.; Aquaro, S. Triclosan: A Small Molecule with Controversial Roles. Antibiotics 2022, 11, 735.
- Shrestha, P.; Zhang, Y.; Chen, W.-J.; Wong, T.-Y. Triclosan: Antimicrobial mechanisms, antibiotics interactions, clinical applications, and human health. J. Environ. Sci. Health Part C 2020, 38, 245–268.
- Heath, R.J.; Rubin, J.R.; Holland, D.R.; Zhang, E.; Snow, M.E.; Rock, C.O. Mechanism of Triclosan Inhibition of Bacterial Fatty Acid Synthesis. J. Biol. Chem. 1999, 274, 11110–11114.
- Abbott, A. Italian scientists under investigation after olive-tree deaths. Nature 2015.
- Poole, A.C.; Pischel, L.; Ley, C.; Suh, G.; Goodrich, J.K.; Haggerty, T.D.; Ley, R.E.; Parsonnet, J. Crossover Control Study of the Effect of Personal Care Products Containing Triclosan on the Microbiome. mSphere 2016, 1, e00056-15.
- Kim, S.A.; Moon, H.; Lee, K.; Rhee, M.S. Bactericidal effects of triclosan in soap both in vitro and in vivo. J. Antimicrob. Chemother. 2015, 70, 3345–3352.
- Ma, Y.; Guo, Y.; Ye, H.; Zhang, J.; Ke, Y. Perinatal Triclosan exposure in the rat induces long-term disturbances in metabolism and gut microbiota in adulthood and old age. Environ. Res. 2019, 182, 109004.
- Giuliano, C.A.; Rybak, M.J. Efficacy of Triclosan as an Antimicrobial Hand Soap and Its Potential Impact on Antimicrobial Resistance: A Focused Review. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2015, 35, 328–336.
- Sun, D.; Zuo, C.; Huang, W.; Wang, J.; Zhang, Z. Triclosan targeting of gut microbiome ameliorates hepatic steatosis in high fat diet-fed mice. J. Antibiot. 2022, 75, 341–353.
- Bever, C.S.; Rand, A.A.; Nording, M.; Taft, D.; Kalanetra, K.M.; Mills, D.A.; Breck, M.A.; Smilowitz, J.T.; German, J.B.; Hammock, B.D. Effects of triclosan in breast milk on the infant fecal microbiome. Chemosphere 2018, 203, 467–473.
- Van Dyke, T.E.; Bartold, P.M.; Reynolds, E.C. The Nexus Between Periodontal Inflammation and Dysbiosis. Front. Immunol. 2020, 11, 511.
- Arimatsu, K.; Yamada, H.; Miyazawa, H.; Minagawa, T.; Nakajima, M.; Ryder, M.I.; Gotoh, K.; Motooka, D.; Nakamura, S.; Iida, T.; et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci. Rep. 2015, 4, 4828.
- Olsen, I.; Yamazaki, K. Can oral bacteria affect the microbiome of the gut? J. Oral Microbiol. 2019, 11, 1586422.
- Komazaki, R.; Katagiri, S.; Takahashi, H.; Maekawa, S.; Shiba, T.; Takeuchi, Y.; Kitajima, Y.; Ohtsu, A.; Udagawa, S.; Sasaki, N.; et al. Periodontal pathogenic bacteria, Aggregatibacter actinomycetemcomitans affect non-alcoholic fatty liver disease by altering gut microbiota and glucose metabolism. Sci. Rep. 2017, 7, 13950.
- Bajaj, J.S.; Matin, P.; White, M.B.; Fagan, A.; Deeb, J.G.; Acharya, C.; Dalmet, S.S.; Sikaroodi, M.; Gillevet, P.M.; Sahingur, S.E. Periodontal therapy favorably modulates the oral-gut-hepatic axis in cirrhosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G824–G837.
- Davies, R.M. The clinical efficacy of triclosan/copolymer and other common therapeutic approaches to periodontal health. Clin. Microbiol. Infect. 2007, 13, 25–29.
- Wallet, M.A.; Calderon, N.L.; Alonso, T.R.; Choe, C.S.; Catalfamo, D.L.; Lalane, C.J.; Neiva, K.G.; Panagakos, F.; Wallet, S.M. Triclosan alters antimicrobial and inflammatory responses of epithelial cells. Oral Dis. 2012, 19, 296–302.
- Meade, K.G.; O’Farrelly, C. β-Defensins: Farming the Microbiome for Homeostasis and Health. Front. Immunol. 2019, 9, 3072.
- Wu, J.-Y.; Chang, M.-C.; Chen, C.-S.; Lin, H.-C.; Tsai, H.-P.; Yang, C.-C.; Yang, C.-H.; Lin, C.-M. Topoisomerase i inhibitor evodiamine acts as an antibacterial agent against drug-resistant Klebsiella pneumoniae. Planta Medica 2012, 79, 27–29.
- Zhu, L.-Q.; Zhang, L.; Zhang, J.; Chang, G.-L.; Liu, G.; Yu, D.-D.; Yu, X.-M.; Zhao, M.-S.; Ye, B. Evodiamine inhibits high-fat diet-induced colitis-associated cancer in mice through regulating the gut microbiota. J. Integr. Med. 2020, 19, 56–65.
- Wang, M.; Zhou, B.; Cong, W.; Zhang, M.; Li, Z.; Li, Y.; Liang, S.; Chen, K.; Yang, D.; Wu, Z. Amelioration of AOM/DSS-Induced Murine Colitis-Associated Cancer by Evodiamine Intervention is Primarily Associated with Gut Microbiota-Metabolism-Inflammatory Signaling Axis. Front. Pharmacol. 2021, 12, 797605.
- Shen, P.; Zhang, Z.; Zhu, K.; Cao, H.; Liu, J.; Lu, X.; Li, Y.; Jing, Y.; Yuan, X.; Fu, Y.; et al. Evodiamine prevents dextran sulfate sodium-induced murine experimental colitis via the regulation of NF-κB and NLRP3 inflammasome. Biomed. Pharmacother. 2018, 110, 786–795.
- Yang, J.Y.; Kim, J.-B.; Lee, P.; Kim, S.-H. Evodiamine Inhibits Helicobacter pylori Growth and Helicobacter pylori-Induced Inflammation. Int. J. Mol. Sci. 2021, 22, 3385.
- Dai, Y.; Zhu, W.; Zhou, J.; Shen, T. The combination of berberine and evodiamine ameliorates high-fat diet-induced non-alcoholic fatty liver disease associated with modulation of gut microbiota in rats. Braz. J. Med. Biol. Res. 2022, 55, e12096.
- Mesas, C.; Martínez, R.; Doello, K.; Ortiz, R.; López-Jurado, M.; Bermúdez, F.; Quiñonero, F.; Prados, J.; Porres, J.; Melguizo, C. In vivo antitumor activity of Euphorbia lathyris ethanol extract in colon cancer models. Biomed. Pharmacother. 2022, 149, 112883.
- Sforcin, J.M. Biological Properties and Therapeutic Applications of Propolis. Phytother. Res. 2016, 30, 894–905.
- Garzarella, E.U.; Navajas-Porras, B.; Pérez-Burillo, S.; Ullah, H.; Esposito, C.; Santarcangelo, C.; Hinojosa-Nogueira, D.; Pastoriza, S.; Zaccaria, V.; Xiao, J.; et al. Evaluating the effects of a standardized polyphenol mixture extracted from poplar-type propolis on healthy and diseased human gut microbiota. Biomed. Pharmacother. 2022, 148, 112759.
- Xue, M.; Liu, Y.; Xu, H.; Zhou, Z.; Ma, Y.; Sun, T.; Liu, M.; Zhang, H.; Liang, H. Propolis modulates the gut microbiota and improves the intestinal mucosal barrier function in diabetic rats. Biomed. Pharmacother. 2019, 118, 109393.
- Efenberger-Szmechtyk, M.; Nowak, A.; Czyzowska, A. Plant extracts rich in polyphenols: Antibacterial agents and natural preservatives for meat and meat products. Crit. Rev. Food Sci. Nutr. 2020, 61, 149–178.
- Chung, K.-T.; Lu, Z.; Chou, M. Mechanism of inhibition of tannic acid and related compounds on the growth of intestinal bacteria. Food Chem. Toxicol. 1998, 36, 1053–1060.
- Zhao, W.-H.; Hu, Z.-Q.; Hara, Y.; Shimamura, T. Inhibition of Penicillinase by Epigallocatechin Gallate Resulting in Restoration of Antibacterial Activity of Penicillin against Penicillinase-Producing Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 2266–2268.
- Yi, S.; Wang, W.; Bai, F.; Zhu, J.; Li, J.; Li, X.; Xu, Y.; Sun, T.; He, Y. Antimicrobial effect and membrane-active mechanism of tea polyphenols against Serratia marcescens. World J. Microbiol. Biotechnol. 2013, 30, 451–460.
- Berman, A.Y.; Motechin, R.A.; Wiesenfeld, M.Y.; Holz, M.K. The therapeutic potential of resveratrol: A review of clinical trials. NPJ Precis. Oncol. 2017, 1, 35.
- Zhao, Y.; Jiang, Q. Roles of the Polyphenol–Gut Microbiota Interaction in Alleviating Colitis and Preventing Colitis-Associated Colorectal Cancer. Adv. Nutr. Int. Rev. J. 2021, 12, 546–565.
- Qi, L.; Liang, R.; Duan, J.; Song, S.; Pan, Y.; Liu, H.; Zhu, M.; Li, L. Synergistic antibacterial and anti-biofilm activities of resveratrol and polymyxin B against multidrug-resistant Pseudomonas aeruginosa. J. Antibiot. 2022, 75, 567–575.
- Ruan, X.; Deng, X.; Tan, M.; Wang, Y.; Hu, J.; Sun, Y.; Yu, C.; Zhang, M.; Jiang, N.; Jiang, R. Effect of resveratrol on the biofilm formation and physiological properties of avian pathogenic Escherichia coli. J. Proteom. 2021, 249, 104357.
- Alrafas, H.R.; Busbee, P.B.; Nagarkatti, M.; Nagarkatti, P.S. Resveratrol modulates the gut microbiota to prevent murine colitis development through induction of Tregs and suppression of Th17 cells. J. Leukoc. Biol. 2019, 106, 467–480.
- Alrafas, H.R.; Busbee, P.B.; Nagarkatti, M.; Nagarkatti, P.S. Resveratrol Downregulates miR-31 to Promote T Regulatory Cells during Prevention of TNBS-Induced Colitis. Mol. Nutr. Food Res. 2019, 64, e1900633.
- Rouas, R.; Kazan, H.F.; El Zein, N.; Lewalle, P.; Rothe, F.; Simion, A.; Akl, H.; Mourtada, M.; El Rifai, M.; Burny, A.; et al. Human natural Treg microRNA signature: Role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur. J. Immunol. 2009, 39, 1608–1618.
- aWang, P.; Wang, J.; Li, D.; Ke, W.; Chen, F.; Hu, X. Targeting the gut microbiota with resveratrol: A demonstration of novel evidence for the management of hepatic steatosis. J. Nutr. Biochem. 2020, 81, 108363.
- Most, J.; Penders, J.; Lucchesi, M.; Goossens, G.H.; Blaak, E.E. Gut microbiota composition in relation to the metabolic response to 12-week combined polyphenol supplementation in overweight men and women. Eur. J. Clin. Nutr. 2017, 71, 1040–1045.
- Konopelski, P.; Mogilnicka, I. Biological Effects of Indole-3-Propionic Acid, a Gut Microbiota-Derived Metabolite, and Its Precursor Tryptophan in Mammals’ Health and Disease. Int. J. Mol. Sci. 2022, 23, 1222.
- Fang, H.; Fang, M.; Wang, Y.; Zhang, H.; Li, J.; Chen, J.; Wu, Q.; He, L.; Xu, J.; Deng, J.; et al. Indole-3-Propionic Acid as a Potential Therapeutic Agent for Sepsis-Induced Gut Microbiota Disturbance. Microbiol. Spectr. 2022, 10, e00125-22.
- Zhao, Z.-H.; Xin, F.-Z.; Xue, Y.; Hu, Z.; Han, Y.; Ma, F.; Zhou, D.; Liu, X.-L.; Cui, A.; Liu, Z.; et al. Indole-3-propionic acid inhibits gut dysbiosis and endotoxin leakage to attenuate steatohepatitis in rats. Exp. Mol. Med. 2019, 51, 1–14.
- Xiao, H.-W.; Cui, M.; Li, Y.; Dong, J.-L.; Zhang, S.-Q.; Zhu, C.-C.; Jiang, M.; Zhu, T.; Wang, B.; Wang, H.-C.; et al. Gut microbiota-derived indole 3-propionic acid protects against radiation toxicity via retaining acyl-CoA-binding protein. Microbiome 2020, 8, 69.
More
Information
Subjects:
Gastroenterology & Hepatology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
995
Revisions:
2 times
(View History)
Update Date:
22 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





