
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Matteo Ardini | -- | 1594 | 2023-05-20 21:01:42 | | | |
| 2 | Peter Tang | Meta information modification | 1594 | 2023-05-22 05:22:02 | | |
Video Upload Options
Affibodies and designed ankyrin repeat proteins (DARPins) are synthetic proteins originally derived from the Staphylococcus aureus virulence factor protein A and the human ankyrin repeat proteins, respectively. The use of these molecules in healthcare has been recently proposed as they are endowed with biochemical and biophysical features heavily demanded to target and fight diseases, as they have a strong binding affinity, solubility, small size, multiple functionalization sites, biocompatibility, and are easy to produce; furthermore, impressive chemical and thermal stability can be achieved, especially when using affibodies. In this sense, several examples reporting on affibodies and DARPins conjugated to nanomaterials have been published, demonstrating their suitability and feasibility in nanomedicine for cancer therapy.
1. Introduction
2. Structural and Biochemical Features of Affibodies and DARPins
2.1. Affibodies
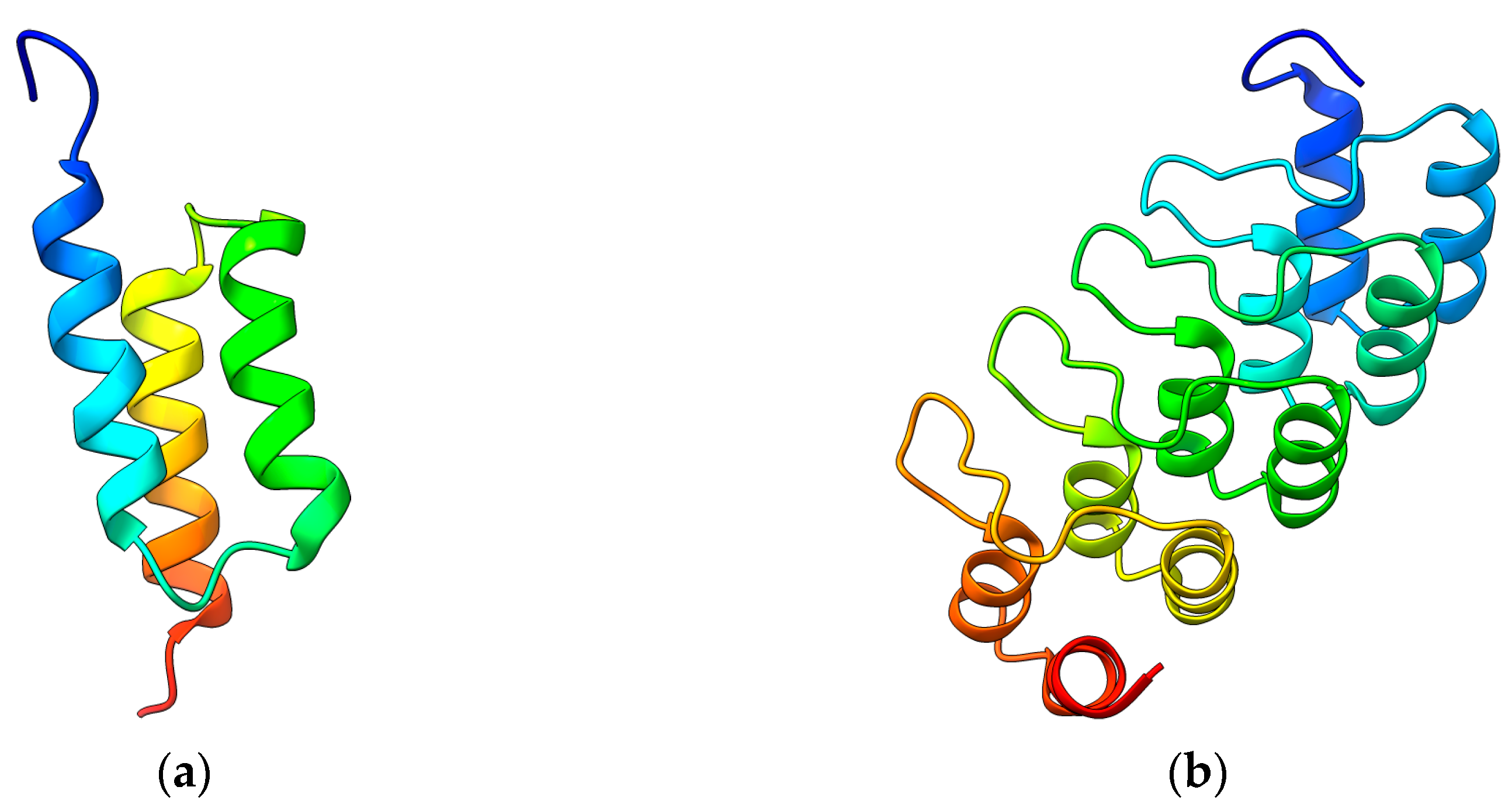
2.2. DARPins
3. Affibody- and DARPin-Conjugated Nanomaterials in Cancer Therapy
A brief description is recalled herein, and a comprehensive overview is provided in Table 1, describing main properties in terms of constitutive matter, shape, and conjugation strategy.
Table 1. Main features of the affibody- and DARPin-conjugated nanomaterials cited here.
|
Inorganic Nanomaterials |
|||||
|---|---|---|---|---|---|
|
Material |
Synthesis |
Shape |
Size 1 |
Bioconjugation Strategy |
Reference |
|
Ag |
Biological synthesis |
Particle |
35 nm |
Crosslinking with EDC/NHS |
[39] |
|
Au |
Chemical synthesis |
Rod |
50 × 8 nm |
Crosslinking with 2-iminothiolane hydrochloride and sulfo-EMCS |
[40] |
|
Ag |
Chemical synthesis |
Particle |
120 nm |
Crosslinking with sulfo-SMCC or EDAC/NHS |
[41] |
|
Au |
Chemical synthesis |
Particle |
31–39 nm |
Crosslinking with sulfo-EMCS |
[42] |
|
Nd, Yb and Tm |
Chemical synthesis |
Particle |
18 nm |
Crosslinking with NHS-PEG-azide |
[43] |
|
Pb, S |
Chemical synthesis |
Dot |
5 nm |
Crosslinking with EDC/NHS |
[44] |
|
Fe3O4, Fe3S4 |
Biological synthesis |
Particle |
30–120 nm |
Crosslinking with SPDP |
[45] |
|
Organic Nanomaterials |
|||||
|
Material |
Synthesis |
Shape |
Size 1 |
Bioconjugation Strategy |
Reference |
|
PLGA |
Chemical synthesis |
Particle |
120 nm |
Crosslinking with EDC/NHS |
[46] |
|
RALA |
Biological synthesis |
Particle |
104.5 nm |
Fusion synthesis |
[47] |
|
MMAE |
Chemical synthesis |
Micelle |
153 nm |
Crosslinking with valine-citrulline dipeptide and PABC spacer |
[48] |
|
MMAE |
Chemical synthesis |
Micelle |
130 nm |
Crosslinking with valine-citrulline dipeptide and PABC spacer |
[49] |
|
PLGA |
Chemical synthesis |
Particle |
218 nm |
Fusion synthesis; protein-protein high affinity interaction |
[50] |
|
PLGA |
Chemical synthesis |
Particle |
140 nm |
Fusion synthesis; crosslinking with EDC/NHS |
[51] |
|
Hybrid Nanomaterials |
|||||
|
Material |
Synthesis |
Shape |
Size 1 |
Bioconjugation Strategy |
Reference |
|
PDA, MnO2 |
Chemical synthesis |
Particle |
163 nm |
Fusion synthesis; crosslinking with Michael addition/Schiff base reaction |
[52] |
|
CaCO3, Fe3O4, polyarginine, dextran sulfate |
Chemical synthesis |
Particle |
400 nm |
Crosslinking with EDC/NHS |
[53] |
|
Biological Nanomaterials |
|||||
|
Material |
Synthesis |
Shape |
Size 1 |
Bioconjugation Strategy |
Reference |
|
Hydrogenated soybean phosphatidylcholine, cholesterol and mPEG 2000-DSPE |
Chemical synthesis |
Micelle |
110–137 nm |
Crosslinking with maleimide-PEG DSPE |
[54] |
|
Hydrogenated soybean phosphatidylcholine, cholesterol, and mPEG 2000-DSPE |
Chemical synthesis |
Micelle |
140 nm |
Crosslinking with maleimide-PEG DSPE |
[55] |
|
L-α-phosphatidylcholine and phosphatidylethanolamine |
Chemical synthesis |
Micelle |
117 nm |
Crosslinking with 2-iminothiolane hydrochloride and sulfo-EMCS |
[56] |
|
AaLS protein |
Biological synthesis |
Particle |
40 nm |
Crosslinking through spontaneous ST-SC isopeptide covalent bond |
[57] |
|
DNA |
Chemical synthesis |
Tetrahedron |
23 nm |
Crosslinking with EMCS |
[58] |
|
DNA |
Chemical synthesis |
Micelle |
132 nm |
Crosslinking with EMCS |
[59] |
1 Average size.
References
- Jin, S.; Sun, Y.; Liang, X.; Gu, X.; Ning, J.; Xu, Y.; Chen, S.; Pan, L. Emerging new therapeutic antibody derivatives for cancer treatment. Sig. Transduct. Target Ther. 2022, 7, 39.
- Gasser, M.; Waaga-Gasser, A.M. Therapeutic Antibodies in Cancer Therapy. Adv. Exp. Med. Biol. 2016, 917, 95–120.
- Ståhl, S.; Gräslund, T.; Eriksson Karlström, A.; Frejd, F.Y.; Nygren, P.Å.; Löfblom, J. Affibody Molecules in Biotechnological and Medical Applications. Trends Biotechnol. 2017, 35, 691–712.
- Farahavar, G.; Abolmaali, S.S.; Gholijani, N.; Nejatollahi, F. Antibody-guided nanomedicines as novel breakthrough therapeutic, diagnostic and theranostic tools. Biomater. Sci. 2019, 7, 4000–4016.
- Simeon, R.; Chen, Z. In vitro-engineered non-antibody protein therapeutics. Protein Cell 2018, 9, 3–14.
- Shilova, O.N.; Deyev, S.M. DARPins: Promising Scaffolds for Theranostics. ActaNaturae 2019, 11, 42–53.
- Shipunova, V.O.; Deyev, S.M. Artificial Scaffold Polypeptides As an Efficient Tool for the Targeted Delivery of Nanostructures In Vitro and In Vivo. ActaNaturae 2022, 14, 54–72.
- Löfblom, J.; Feldwisch, J.; Tolmachev, V.; Carlsson, J.; Ståhl, S.; Frejd, F.Y. Affibody molecules: Engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 2010, 584, 2670–2680.
- De, A.; Kuppusamy, G.; Karri, V.V.S.R. Affibody molecules for molecular imaging and targeted drug delivery in the management of breast cancer. Int. J. Biol. Macromol. 2018, 107, 906–919.
- Jia, D.; Liu, H.; Zheng, S.; Yuan, D.; Sun, R.; Wang, F.; Li, Y.; Li, H.; Yuan, F.; Fan, Q.; et al. ICG-Dimeric Her2-Specific Affibody Conjugates for Tumor Imaging and Photothermal Therapy for Her2-Positive Tumors. Mol. Pharm. 2023, 20, 427–437.
- Oroujeni, M.; Bezverkhniaia, E.A.; Xu, T.; Liu, Y.; Plotnikov, E.V.; Karlberg, I.; Ryer, E.; Orlova, A.; Tolmachev, V.; Frejd, F.Y. Evaluation of an Affibody-Based Binder for Imaging of Immune Check-Point Molecule B7-H3. Pharmaceutics 2022, 14, 1780.
- Karsten, L.; Janson, N.; Le Joncour, V.; Alam, S.; Müller, B.; Tanjore Ramanathan, J.; Laakkonen, P.; Sewald, N.; Müller, K.M. Bivalent EGFR-Targeting DARPin-MMAE Conjugates. Int. J. Mol. Sci. 2022, 23, 2468.
- Vorobyeva, A.; Schulga, A.; Rinne, S.S.; Günther, T.; Orlova, A.; Deyev, S.; Tolmachev, V. Indirect Radioiodination of DARPin G3 Using N-succinimidyl-Para-Iodobenzoate Improves the Contrast of HER2 Molecular Imaging. Int. J. Mol. Sci. 2019, 20, 3047.
- Qi, S.; Hoppmann, S.; Xu, Y.; Cheng, Z. PET Imaging of HER2-Positive Tumors with Cu-64-Labeled Affibody Molecules. Mol. Imaging Biol. 2019, 21, 907–916.
- Shipunova, V.O.; Kolesnikova, O.A.; Kotelnikova, P.A.; Soloviev, V.D.; Popov, A.A.; Proshkina, G.M.; Nikitin, M.P.; Deyev, S.M. Comparative Evaluation of Engineered Polypeptide Scaffolds in HER2-Targeting Magnetic Nanocarrier Delivery. ACS Omega 2021, 10, 16000–16008.
- Uhlén, M.; Guss, B.; Nilsson, B.; Gatenbeck, S.; Philipson, L.; Lindberg, M. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J. Biol. Chem. 1984, 259, 1695–1702.
- Moks, T.; Abrahmsén, L.; Nilsson, B.; Hellman, U.; Sjöquist, J.; Uhlén, M. Staphylococcal protein A consists of five IgG-binding domains. Eur. J. Biochem. 1986, 156, 637–643.
- Ibrahim, S. Immunoglobulin Binding Specificities of the Homology Regions (Domains) of Protein A. Scand. J. Immunol. 1993, 38, 368–374.
- Jansson, B.; Uhlén, M.; Nygren, P.-Å. All individual domains of staphylococcal protein A show Fab binding. FEMS Immunol. Med. Microbiol. 1998, 20, 69–78.
- Gouda, H.; Torigoe, H.; Saito, A.; Sato, M.; Arata, Y.; Shimada, I. Three-dimensional solution structure of the B domain of staphylococcal protein A: Comparisons of the solution and crystal structures. Biochemistry 1992, 31, 9665–9672.
- Starovasnik, M.A.; Skelton, N.J.; O’Connell, M.P.; Kelley, R.F.; Reilly, D.; Fairbrother, W.J. Solution Structure of the E-Domain of Staphylococcal Protein A. Biochemistry 1996, 35, 15558–15569.
- Nilsson, B.; Moks, T.; Jansson, B.; Abrahmsén, L.; Elmblad, A.; Holmgren, E.; Henrichson, C.; Jones, T.A.; Uhlén, M. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. Des. Sel. 1987, 1, 107–113.
- Tashiro, M.; Tejero, R.; Zimmerman, D.E.; Celda, B.; Nilsson, B.; Montelione, G.T. High-resolution solution NMR structure of the Z domain of staphylococcal protein A. J. Mol. Biol. 1997, 272, 573–590.
- Zheng, D.; Aramini, J.M.; Montelione, G.T. Validation of helical tilt angles in the solution NMR structure of the Z domain of Staphylococcal protein A by combined analysis of residual dipolar coupling and NOE data. Protein Sci. 2004, 13, 549–554.
- Nord, K.; Nilsson, J.; Nilsson, B.; Uhlén, M.; Nygren, P.A. A combinatorial library of an alpha-helical bacterial receptor domain. Protein Eng. Des. Sel. 1995, 8, 601–608.
- Nord, K.; Gunneriusson, E.; Ringdahl, J.; Ståhl, S.; Uhlén, M.; Nygren, P. Binding proteins selected from combinatorial libraries of an α-helical bacterial receptor domain. Nat. Biotechnol. 1997, 15, 772–777.
- Feldwisch, J.; Tolmachev, V.; Lendel, C.; Herne, N.; Sjöberg, A.; Larsson, B.; Rosik, D.; Lindqvist, E.; Fant, G.; Höidén-Guthenberg, I.; et al. Design of an optimized scaffold for affibody molecules. J. Mol. Biol. 2010, 398, 232–247.
- Votsmeier, C.; Plittersdorf, H.; Hesse, O.; Scheidig, A.; Strerath, M.; Gritzan, U.; Pellengahr, K.; Scholz, P.; Eicker, A.; Myszka, D.; et al. Femtomolar Fab binding affinities to a protein target by alternative CDR residue co-optimization strategies without phage or cell surface display. mAbs 2012, 4, 341–348.
- Arora, P.; Oas, T.G.; Myers, J.K. Fast and faster: A designed variant of the B-domain of protein A folds in 3 microsec. Protein Sci. 2004, 13, 847–853.
- Eigenbrot, C.; Ultsch, M.; Dubnovitsky, A.; Abrahmsén, L.; Härd, T. Structural basis for high-affinity HER2 receptor binding by an engineered protein. Proc. Natl. Acad. Sci. USA 2010, 107, 15039–15044.
- Merz, T.; Wetzel, S.K.; Firbank, S.; Plückthun, A.; Grütter, M.G.; Mittl, P.R. Stabilizing ionic interactions in a full-consensus ankyrin repeat protein. J. Mol. Biol. 2008, 376, 232–240.
- Breeden, L.; Nasmyth, K. Similarity between cell-cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila. Nature 1987, 329, 651–654.
- Kane, E.I.; Spratt, D.E. Structural Insights into Ankyrin Repeat-Containing Proteins and Their Influence in Ubiquitylation. Int. J. Mol. Sci. 2021, 22, 609.
- Binz, H.K.; Stumpp, M.T.; Forrer, P.; Amstutz, P.; Plückthun, A. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 2003, 332, 489–503.
- Steiner, D.; Forrer, P.; Plückthun, A. Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. J. Mol. Biol. 2008, 382, 1211–1227.
- Schilling, J.; Schoeppe, J.; Plückthun, A. From DARPins to LoopDARPins: Novel LoopDARPin design allows the selection of low picomolar binders in a single round of ribosome display. J. Mol. Biol. 2014, 426, 691–721.
- Schilling, J.; Jost, C.; Ilie, I.M.; Schnabl, J.; Buechi, O.; Eapen, R.S.; Truffer, R.; Caflisch, A.; Forrer, P. Thermostable designed ankyrin repeat proteins (DARPins) as building blocks for innovative drugs. J. Biol. Chem. 2022, 298, 101403.
- Plückthun, A. Designed ankyrin repeat proteins (DARPins): Binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 489–511.
- Shipunova, V.O.; Belova, M.M.; Kotelnikova, P.A.; Shilova, O.N.; Mirkasymov, A.B.; Danilova, N.V.; Komedchikova, E.N.; Popovtzer, R.; Deyev, S.M.; Nikitin, M.P. Photothermal Therapy with HER2-Targeted Silver Nanoparticles Leading to Cancer Remission. Pharmaceutics 2022, 14, 1013.
- Proshkina, G.M.; Shramova, E.I.; Shilova, M.V.; Zelepukin, I.V.; Shipunova, V.O.; Ryabova, A.V.; Deyev, S.M.; Kotlyar, A.B. DARPin_9-29-Targeted Gold Nanorods Selectively Suppress HER2-Positive Tumor Growth in Mice. Cancers 2021, 13, 5235.
- Pourshohod, A.; Zeinali, M.; Ghaffari, M.A.; Kheirollah, A.; Jamalan, M. Improvement of specific aiming of X-ray radiotherapy on HER2-overexpressing cancerous cell lines by targeted delivery of silver nanoparticle. J. Drug Delivery Sci. Technol. 2022, 76, 103746.
- Zhang, C.; Zhang, F.; Han, M.; Wang, X.; Du, J.; Zhang, H.; Li, W. Co-delivery of 5-fluorodeoxyuridine and doxorubicin via gold nanoparticle equipped with affibody-DNA hybrid strands for targeted synergistic chemotherapy of HER2 overexpressing breast cancer. Sci. Rep. 2020, 10, 22015.
- Roy, S.; Curry, S.D.; Corbella Bagot, C.; Mueller, E.N.; Mansouri, A.M.; Park, W.; Cha, J.N.; Goodwin, A.P. Enzyme Prodrug Therapy with Photo-Cross-Linkable Anti-EGFR Affibodies Conjugated to Upconverting Nanoparticles. ACS Nano 2022, 16, 15873–15883.
- Al-Ani, A.W.; Zamberlan, F.; Ferreira, L.; Bradshaw, T.D.; Thomas, N.R.; Turyanska, L. Near-infrared PbS quantum dots functionalized with affibodies and ZnPP for targeted imaging and therapeutic applications. Nano Express 2021, 2, 040005.
- Ma, S.; Gu, C.; Xu, J.; He, J.; Li, S.; Zheng, H.; Pang, B.; Wen, Y.; Fang, Q.; Liu, W.; et al. Strategy for Avoiding Protein Corona Inhibition of Targeted Drug Delivery by Linking Recombinant Affibody Scaffold to Magnetosomes. Int. J. Nanomed. 2022, 17, 665–680.
- Shipunova, V.O.; Sogomonyan, A.S.; Zelepukin, I.V.; Nikitin, M.P.; Deyev, S.M. PLGA Nanoparticles Decorated with Anti-HER2 Affibody for Targeted Delivery and Photoinduced Cell Death. Molecules 2021, 26, 3955.
- Zhang, F.; Yin, J.; Zhang, C.; Han, M.; Wang, X.; Fu, S.; Du, J.; Zhang, H.; Li, W. Affibody-Conjugated RALA Polymers Delivering Oligomeric 5-Fluorodeoxyuridine for Targeted Therapy of HER2 Overexpressing Gastric Cancer. Macromol. Biosci. 2020, 20, e2000083.
- Xia, X.; Yang, X.; Huang, W.; Xia, X.; Yan, D. Self-Assembled Nanomicelles of Affibody-Drug Conjugate with Excellent Therapeutic Property to Cure Ovary and Breast Cancers. Nano-Micro Lett. 2022, 14, 33.
- Yang, X.; Xia, X.; Huang, W.; Xia, X.; Yan, D. Highly efficient tumor-targeted nanomedicine assembled from affibody-drug conjugate for colorectal cancer therapy. Nano Res. 2023, 16, 5256–5264.
- Komedchikova, E.N.; Kolesnikova, O.A.; Tereshina, E.D.; Kotelnikova, P.A.; Sogomonyan, A.S.; Stepanov, A.V.; Deyev, S.M.; Nikitin, M.P.; Shipunova, V.O. Two-Step Targeted Drug Delivery via Proteinaceous Barnase-Barstar Interface and Doxorubicin-Loaded Nano-PLGA Outperforms One-Step Strategy for Targeted Delivery to HER2-Overexpressing Cells. Pharmaceutics 2023, 15, 52.
- Shipunova, V.O.; Komedchikova, E.N.; Kotelnikova, P.A.; Zelepukin, I.V.; Schulga, A.A.; Proshkina, G.M.; Shramova, E.I.; Kutscher, H.L.; Telegin, G.B.; Kabashin, A.V.; et al. Dual Regioselective Targeting the Same Receptor in Nanoparticle-Mediated Combination Immuno/Chemotherapy for Enhanced Image-Guided Cancer Treatment. ACS Nano 2020, 27, 12781–12795.
- Wang, H.; Jia, D.; Yuan, D.; Yin, X.; Yuan, F.; Wang, F.; Shi, W.; Li, H.; Zhu, L.-M.; Fan, Q. Dimeric Her2-specific affibody mediated cisplatin-loaded nanoparticles for tumor enhanced chemo-radiotherapy. J. Nanobiotechnol. 2021, 19, 138.
- Novoselova, M.V.; Shramova, E.I.; Sergeeva, O.V.; Shcherbinina, E.Y.; Perevoschikov, S.V.; Melnikov, P.; Griaznova, O.Y.; Sergeev, I.S.; Konovalova, E.V.; Schulga, A.A.; et al. Polymer/magnetite carriers functionalized by HER2-DARPin: Avoiding lysosomes during internalization and controlled toxicity of doxorubicin by focused ultrasound induced release. Nanomedicine 2023, 47, 102612.
- Jia, D.; Yang, Y.; Yuan, F.; Fan, Q.; Wang, F.; Huang, Y.; Song, H.; Hu, P.; Wang, R.; Li, G.; et al. Increasing the antitumor efficacy of doxorubicin liposomes with coupling an anti-EGFR affibody in EGFR-expressing tumor models. Int. J. Pharm. 2020, 586, 119541.
- Jia, D.; Wang, F.; Yang, Y.; Hu, P.; Song, H.; Lu, Y.; Wang, R.; Li, G.; Liu, R.; Li, J.; et al. Coupling EGFR-Antagonistic Affibody Enhanced Therapeutic Effects of Cisplatin Liposomes in EGFR-expressing Tumor Models. J. Pharm. Sci. 2022, 111, 450–457.
- Shipunova, V.O.; Shramova, E.I.; Schulga, A.A.; Shilova, M.V.; Deyev, S.M.; Proshkina, G.M. Delivery of Barnase to Cells in Liposomes Functionalized by Her2-Specific DARPin Module. Russ. J. Bioorg. Chem. 2020, 46, 1156–1161.
- Jun, H.; Jang, E.; Kim, H.; Yeo, M.; Park, S.G.; Lee, J.; Shin, K.J.; Chae, Y.C.; Kang, S.; Kim, E. TRAIL & EGFR affibody dual-display on a protein nanoparticle synergistically suppresses tumor growth. J. Control Release 2022, 349, 367–378.
- Zhang, C.; Han, M.; Zhang, F.; Yang, X.; Du, J.; Zhang, H.; Li, W.; Chen, S. Enhancing Antitumor Efficacy of Nucleoside Analog 5-Fluorodeoxyuridine on HER2-Overexpressing Breast Cancer by Affibody-Engineered DNA Nanoparticle. Int. J. Nanomed. 2020, 15, 885–900.
- Zhang, C.; Fu, S.; Zhang, F.; Han, M.; Wang, X.; Du, J.; Zhang, H.; Li, W. Affibody Modified G-quadruplex DNA Micelles Incorporating Polymeric 5-Fluorodeoxyuridine for Targeted Delivery of Curcumin to Enhance Synergetic Therapy of HER2 Positive Gastric Cancer. Nanomaterials 2022, 12, 696.




