
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Andrei Surguchov | -- | 2134 | 2023-05-10 00:03:26 | | | |
| 2 | Rita Xu | Meta information modification | 2134 | 2023-05-10 03:57:47 | | |
Video Upload Options
Synucleinopathies are a group of neurodegenerative diseases with common pathological lesions associated with the excessive accumulation and abnormal intracellular deposition of toxic species of α-synuclein. The shared clinical features are chronic progressive decline of motor, cognitive, and behavioral functions. These disorders include Parkinson’s disease, dementia with Lewy body, and multiple system atrophy. Vigorous research in the mechanisms of pathology of these illnesses is currently under way to find disease-modifying treatment and molecular markers for early diagnosis. α-Synuclein is a prone-to-aggregate, small amyloidogenic protein with multiple roles in synaptic vesicle trafficking, neurotransmitter release, and intracellular signaling events. Its expression is controlled by several mechanisms, one of which is epigenetic regulation.
1. Introduction
2. Epigenetic Regulation of α-Synuclein
3. α-Synuclein as a Genetic Modulator
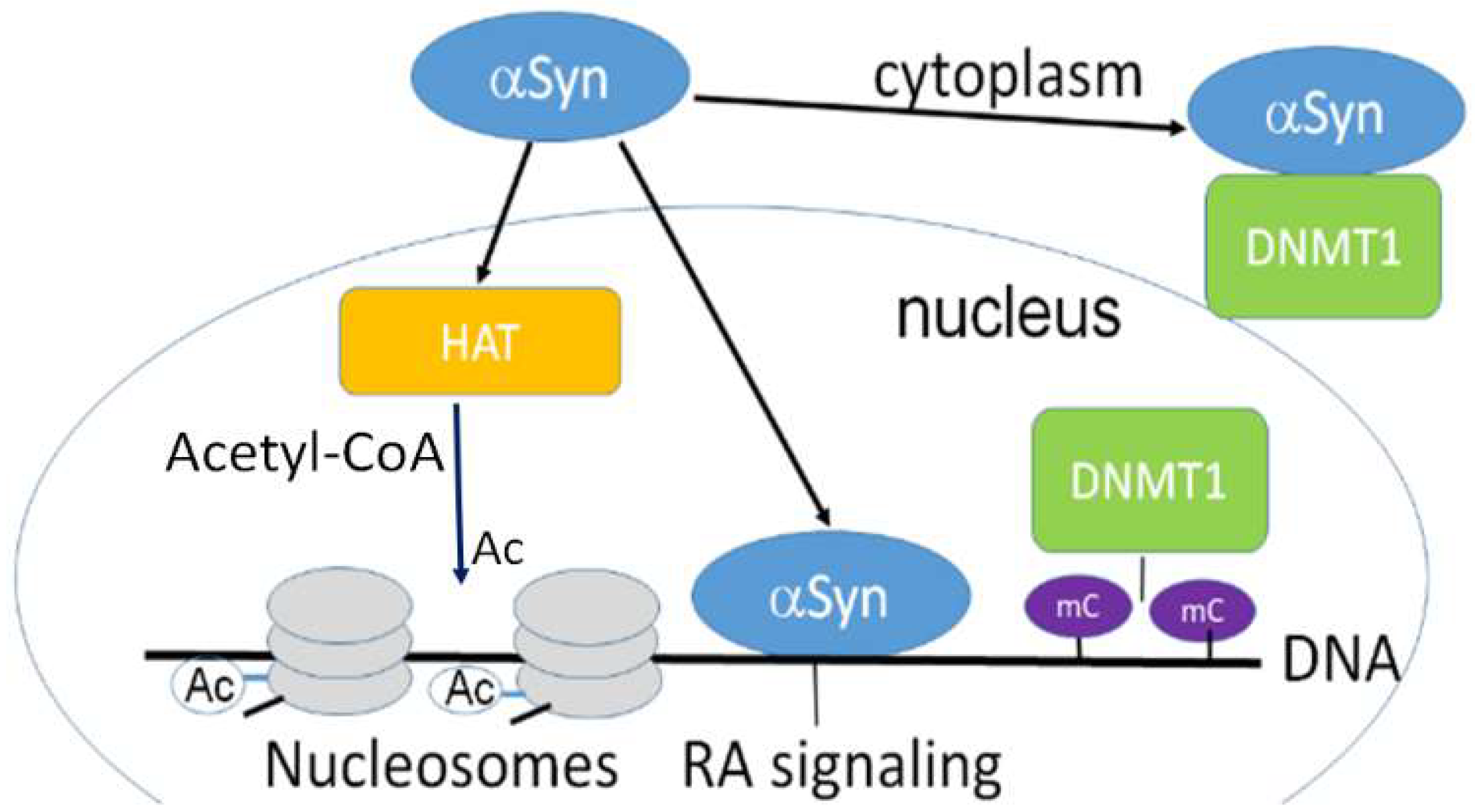
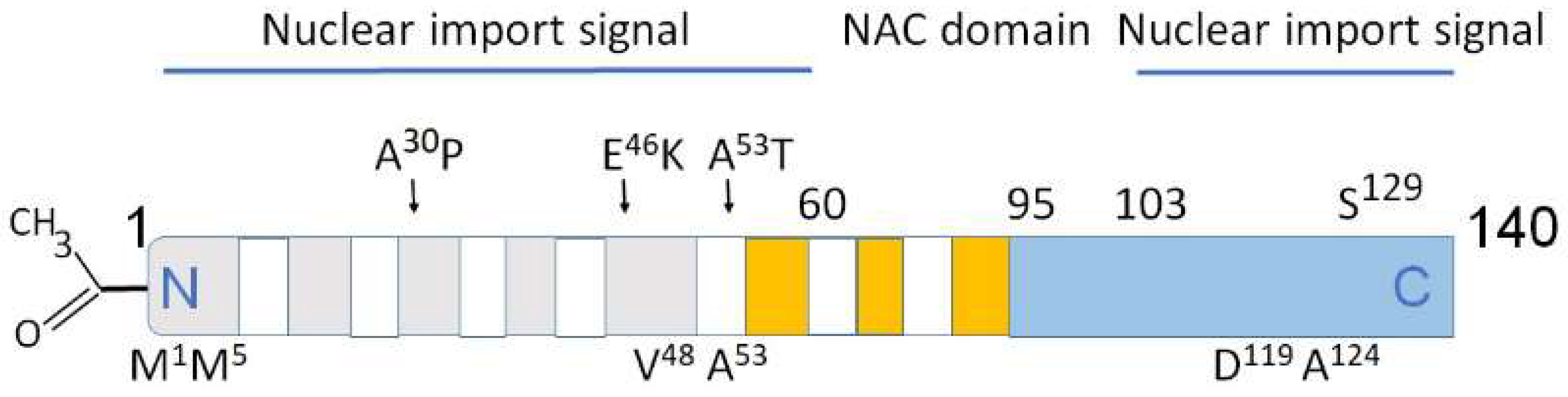
3.1. Nuclear Localization of α-Synuclein and DNA Binding
3.2. Nuclear α-Synuclein Regulates Histone Modifications
References
- Park, J.; Lee, K.; Kim, K.; Yi, S.-J. The role of histone modifications: From neurodevelopment to neurodiseases. Signal Transduct. Target. Ther. 2022, 7, 217.
- Miranda-Morales, E.; Meier, K.; Sandoval-Carrillo, A.; Salas-Pacheco, J.; Vázquez-Cárdenas, P.; Arias-Carrión, O. Implications of DNA methylation in parkinson’s disease. Front. Mol. NeuroSci. 2017, 10, 225.
- Rathore, A.S.; Birla, H.; Singh, S.S.; Zahra, W.; Dilnashin, H.; Singh, R.; Keshri, P.K.; Singh, S.P. Epigenetic Modulation in Parkinson’s Disease and Potential Treatment Therapies. Neurochem. Res. 2021, 46, 1618–1626.
- Surguchov, A. Biomarkers in Parkinson’s Disease. In Neuro degenerative Diseases Biomarkers; Part of the Neuromethods book series; Peplow, P.V., Martinez, B., Gennarelli, T.A., Eds.; Humana: New York, NY, USA, 2022; Volume 173, pp. 155–180.
- Schaffner, S.L.; Kobor, M.S. DNA methylation as a mediator of genetic and environmental influences on Parkinson’s disease susceptibility: Impacts of alpha-Synuclein, physical activity, and pesticide exposure on the epigenome. Front. Genet. 2022, 13, 971298.
- Surguchov, A.; Bernal, L.; Surguchev, A.A. Phytochemicals as Regulators of Genes Involved in Synucleinopathies. Biomolecules 2021, 11, 624.
- Waddington, C.H. Canalization of development and the inheritance of acquired characters. Nature 1942, 150, 563–565.
- Edgar, R.D.; Jones, M.J.; Meaney, M.J.; Turecki, G.; Kobor, M.S. BECon: A tool for interpreting DNA methylation findings from blood in the context of brain. Transl. Psychiatry 2017, 7, e1187.
- Matsumoto, L.; Takuma, H.; Tamaoka, A.; Kurisaki, H.; Date, H.; Tsuji, S.; Iwata, A. CpG Demethylation Enhances Alpha-Synuclein Expression and Affects the Pathogenesis of Parkinson’s Disease. PLoS ONE 2010, 5, e15522.
- Wüllner, U.; Kaut, O.; Deboni, L.; Piston, D.; Schmitt, I. DNA methylation in Parkinson’s disease. J. Neurochem. 2016, 139, 108–120.
- Daniele, S.; Costa, B.; Pietrobono, D.; Giacomelli, C.; Iofrida, C.; Trincavelli, M.L.; Fusi, J.; Franzoni, F.; Martini, C. Epigenetic Modifications of the α-Synuclein Gene and Relative Protein Content Are Affected by Ageing and Physical Exercise in Blood from Healthy Subjects. Oxidative Med. Cell. Longev. 2018, 2018, 3740345.
- Desplats, P.; Spencer, B.; Coffee, E.; Patel, P.; Michael, S.; Patrick, C.; Adame, A.; Rockenstein, E.; Masliah, E. α-Synuclein Sequesters Dnmt1 from the Nucleus. J. Biol. Chem. 2011, 286, 9031–9037.
- Schaffner, S.L.; Wassouf, Z.; Lazaro, D.F.; Xylaki, M.; Gladish, N.; Lin, D.T.S.; MacIsaac, J.; Ramadori, K.; Hentrich, T.; Schulze-Hentrich, J.M.; et al. Alpha-synuclein overexpression induces epigenomic dysregulation of glutamate signaling and locomotor pathways. Hum. Mol. Genet. 2022, 31, 3694–3714.
- Ai, S.-X.; Xu, Q.; Hu, Y.-C.; Song, C.-Y.; Guo, J.-F.; Shen, L.; Wang, C.-R.; Yu, R.-L.; Yan, X.-X.; Tang, B.-S. Hypomethylation of SNCA in blood of patients with sporadic Parkinson’s disease. J. Neurol. Sci. 2014, 337, 123–128.
- Pavlou, M.A.S.; Outeiro, T.F. Epigenetics in Parkinson’s Disease. Neuroepigenomics Aging Dis. 2017, 978, 363–390.
- Pihlstrøm, L.; Berge, V.; Rengmark, A.; Toft, M. Parkinson’s disease correlates with promoter methylation in the α-synuclein gene. Mov. Disord. 2015, 30, 577–580.
- Angelopoulou, E.; Paudel, Y.N.; Papageorgiou, S.G.; Piperi, C. Environmental Impact on the Epigenetic Mechanisms Underlying Parkinson’s Disease Pathogenesis: A Narrative Review. Brain Sci. 2022, 12, 175.
- Yoon, B.-H.; Kim, M.; Kim, M.-H.; Kim, H.-J.; Kim, J.-H.; Kim, J.H.; Kim, J.; Kim, Y.S.; Lee, D.; Kang, S.-J.; et al. Dynamic Transcriptome, DNA Methylome, and DNA Hydroxymethylome Networks During T-Cell Lineage Commitment. Mol. Cells 2018, 41, 953–963.
- Maroteaux, L.; Campanelli, J.T.; Scheller, R.H. Synuclein: A neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J. Neurosci. 1988, 8, 2804–2815.
- Cherny, D.; Hoyer, W.; Subramaniam, V.; Jovin, T.M. Double-stranded DNA Stimulates the Fibrillation of α-Synuclein in vitro and is Associated with the Mature Fibrils: An Electron Microscopy Study. J. Mol. Biol. 2004, 344, 929–938.
- Hegde, M.L.; Rao, K.S. DNA induces folding in alpha-synuclein: Understanding the mechanism using chaperone property of osmolytes. Arch. Biochem. Biophys. 2007, 464, 57–69.
- Muralidhar, L.H.; Hegde, M.L.; Vasudevaraju, P.; Rao, K.J. DNA induced folding/fibrillation of alpha-synuclein: New insights in Parkinson’s disease. Front. Biosci. 2010, 15, 418–436.
- Rao, K.S.; Guerrero, E.; Collen, T.B.; Vasudevaraju, P.; Hegde, M.L.; Britton, G.B. New evidence on α-synuclein and Tau binding to conformation and sequence specific GCFNx01 rich DNA: Relevance to neurological disorders. J. Pharm. Bioallied Sci. 2012, 4, 112–117.
- Schaser, A.J.; Osterberg, V.R.; Dent, S.E.; Stackhouse, T.L.; Wakeham, C.M.; Boutros, S.W.; Weston, L.J.; Owen, N.; Weissman, T.A.; Luna, E.; et al. Alpha-synuclein is a DNA binding protein that modulates DNA repair with implications for Lewy body disorders. Sci. Rep. 2019, 9, 1–19.
- Popova, B.; Wang, D.; Patz, C.; Akkermann, D.; Lazaro, D.F.; Galka, D.; Kolog Gulko, M.; Bohnsack, M.T.; Mobius, W.; Bohnsack, K.E.; et al. DEAD-box RNA helicase Dbp4/DDX10 is an enhancer of α-synuclein toxicity and oligomerization. PLoS Genet. 2021, 17, e1009407.
- Dent, S.E.; King, D.P.; Osterberg, V.R.; Adams, E.K.; Mackiewicz, M.R.; Weissman, T.A.; Unni, V.K. Phosphorylation of the aggregate-forming protein alpha-synuclein on serine-129 inhibits its DNA-bending properties. J. Biol. Chem. 2021, 298, 101552.
- Chung, C.Y.; Khurana, V.; Yi, S.; Sahni, N.; Loh, K.H.; Auluck, P.K.; Baru, V.; Udeshi, N.D.; Freyzon, Y.; Carr, S.A.; et al. In Situ Peroxidase Labeling and Mass-Spectrometry Connects Alpha-Synuclein Directly to Endocytic Trafficking and mRNA Metabolism in Neurons. Cell Syst. 2017, 4, 242–250.e4.
- Kim, S.; Park, J.M.; Moon, J.; Choi, H.J. Alpha-synuclein interferes with cAMP/PKA-dependent upregulation of dopamine β-hydroxylase and is associated with abnormal adaptive responses to immobilization stress. Exp. Neurol. 2014, 252, 63–74.
- Jiang, K.; Rocha, S.; Kumar, R.; Westerlund, F.; Wittung-Stafshede, P. C-terminal truncation of α-synuclein alters DNA structure from extension to compaction. Biochem. Biophys. Res. Commun. 2021, 568, 43–47.
- Kontopoulos, E.; Parvin, J.D.; Feany, M.B. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum. Mol. Genet. 2006, 15, 3012–3023.
- Pinho, R.; Paiva, I.; Jerčić, K.G.; Fonseca-Ornelas, L.; Gerhardt, E.; Fahlbusch, C.; Esparcia, P.G.; Kerimoglu, C.; Pavlou, M.A.S.; Villar-Piqué, A.; et al. Nuclear localization and phosphorylation modulate pathological effects of alpha-synuclein. Hum. Mol. Genet. 2019, 28, 31–50.
- Sharma A, Liu H, Tobar-Tosse F, Noll A, Dakal TC, Li H, Holz FG, Loeffler K, Herwig-Carl MC Genome organization in proximity to the BAP1 locus appears to play a pivotal role in a variety of cancers. Cancer Sci. 2020, 111, 1385–1391.
- Goers, J.; Manning-Bog, A.B.; McCormack, A.L.; Millett, I.S.; Doniach, S.; Di Monte, D.A.; Uversky, V.N.; Fink, A.L. Nuclear Localization of alpha-Synuclein and Its Interaction with Histones. Biochemistry 2003, 42, 8465–8471.
- Sugeno, N.; Jäckel, S.; Voigt, A.; Wassouf, Z.; Schulze-Hentrich, J.; Kahle, P.J. alpha-Synuclein enhances histone H3 lysine-9 dimethylation and H3K9me2-dependent transcriptional responses. Sci. Rep. 2016, 6, 36328.
- Takahashi-Fujigasaki, J.; Fujigasaki, H. Histone deacetylase (HDAC) 4 involvement in both Lewy and Marinesco bodies. Neuropathol. Appl. Neurobiol. 2006, 32, 562–566.
- Wu, Q.; Yang, X.; Zhang, L.; Zhang, Y.; Feng, L. Nuclear Accumulation of Histone Deacetylase 4 (HDAC4) Exerts Neurotoxicity in Models of Parkinson’s Disease. Mol. Neurobiol. 2017, 54, 6970–6983.




