Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Annalisa Abdel Azim | -- | 3592 | 2023-04-30 15:36:44 | | | |
| 2 | Jessie Wu | -8 word(s) | 3584 | 2023-05-04 04:27:37 | | | | |
| 3 | Jessie Wu | Meta information modification | 3584 | 2023-05-04 04:28:14 | | | | |
| 4 | Jessie Wu | Meta information modification | 3584 | 2023-05-04 04:28:56 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Abdel Azim, A.; Bellini, R.; Vizzarro, A.; Bassani, I.; Pirri, C.F.; Menin, B. Biorecovery of Critical Raw Materials through Archaeal factory. Encyclopedia. Available online: https://encyclopedia.pub/entry/43651 (accessed on 07 February 2026).
Abdel Azim A, Bellini R, Vizzarro A, Bassani I, Pirri CF, Menin B. Biorecovery of Critical Raw Materials through Archaeal factory. Encyclopedia. Available at: https://encyclopedia.pub/entry/43651. Accessed February 07, 2026.
Abdel Azim, Annalisa, Ruggero Bellini, Arianna Vizzarro, Ilaria Bassani, Candido Fabrizio Pirri, Barbara Menin. "Biorecovery of Critical Raw Materials through Archaeal factory" Encyclopedia, https://encyclopedia.pub/entry/43651 (accessed February 07, 2026).
Abdel Azim, A., Bellini, R., Vizzarro, A., Bassani, I., Pirri, C.F., & Menin, B. (2023, April 30). Biorecovery of Critical Raw Materials through Archaeal factory. In Encyclopedia. https://encyclopedia.pub/entry/43651
Abdel Azim, Annalisa, et al. "Biorecovery of Critical Raw Materials through Archaeal factory." Encyclopedia. Web. 30 April, 2023.
Copy Citation
Bio-metallurgy is a promising alternative for e-waste valorisation based on biological routes of specialised microorganisms able to leach solid-containing metals. Because of the physiology of these microorganisms, microbial leaching can be economically feasible, besides being an environmentally sustainable process. Like Bacteria and Fungi, Archaea are also capable of metal leaching activity, though their potential is underestimated. Because of the physiology of these microorganisms, microbial leaching can be both economically and environmentally sustainable. Archaea, Bacteria and Fungi, are capable of metal leaching activity, although their potential is underappreciated.
e-waste
archaea
critical raw materials
bioleaching
biomining
critical metals
1. Introduction
The metallurgy process for metal refinement frequently requires energy-intensive melting steps based on the use of fossil fuel. Pyrometallurgy, hydrometallurgy and biohydrometallurgy are three variants of metallurgy that differ in the mechanism of extraction. While pyrometallurgy uses heat power, hydrometallurgy applies redox chemical reactions in an aqueous or organic liquid solution. Biohydrometallurgy works similarly to hydrometallurgical processes, except that the reagents are directly supplied by the microorganisms because of the by-products of their metabolic reactions. Biohydrometallurgy operates similarly to hydrometallurgical processes, except that the chemicals are supplied directly by microorganisms as byproducts of their metabolic reactions. Research is rapidly evolving to manage the challenges in recycling e-waste for metal recovery through both chemical and biological routes. As part of the chemical method, hydrometallurgy is a well-established process for metal leaching from both primary and secondary resources [1][2]. Conversely, biohydrometallurgy is still under investigation in several aspects, such as the physiology of less explored microorganisms, bioprocess operation and scalability [3][4][5].
The biological route is carried out by specialised leaching microorganisms which implement strategies for recovering metals against their scarcity in the environment.
The microbial systems acquire metals necessary required for metabolism and counteract the adverse effects of toxic metals to protect the cell by using a whole repertoire of mechanisms and in order to acclimatise themselves to hostile environmental conditions [6]. Microbial growth, metabolism and differentiation are intimately linked to the biogeochemical cycle of metals. The Metals can be classified into three categories based on their different physiological roles: (a) vital and non-toxic, such as Ca and Mg; (b) vital but toxic at elevated levels, such as Fe, Mn, Zn, Cu, Co, Ni, and Mo; and (c) toxic, such as Hg and Cd [7][8]. Metals such as aluminium (Al), antinomy (Sb), arsenic (As), barium (Ba), beryllium (Be), bismuth (Bi), cadmium (Cd), gallium (Ga), germanium (Ge), gold (Au), indium (In), lead (Pb), lithium (Li), mercury (Hg), nickel (Ni), platinum (Pt), silver (Ag), strontium (Sr), tellurium (Te), thallium (Tl), tin (Sn), titanium (Ti), vanadium (V), and uranium (U) have no established biological functions and are considered as non-essential metals [6]. Nevertheless, there is evidence of microbial removal of some of these non-essential metals [3][4][5][9].
Natural sources of metal-leaching microorganisms are mine sites or acid mine drainage samples, though other sources exist (e.g., brines and sediments, sludge from anaerobic digestion plants) [10]. Due to the physiology of these microorganisms, bioleaching can be economically feasible (e.g., in situ processing is viable thanks to the simplicity of technology design and operating conditions; the possibility of reusing existing facilities; and reasonable capital and operating costs) and eco-friendly approach with higher efficacy, safety and ease of control [11]. Furthermore, the waste streams, including toxic gas emissions and wastewater, are more restricted and controlled in the context of microbial leaching. However, reaction kinetics can be pointed out as the main limitation of biohydrometallurgy processes, being strictly dependent on the characteristics of the biocatalyst.
Factors affecting the bioleaching process include pH, temperature, oxygen, and carbon dioxide supply, as well as nutrients in the medium. pH is selective only for certain metal compounds (e.g., carbonates, common oxides, acid-soluble sulphides), and of course, it is also a filter for microbial growth and activity [12][13][14].
Bioleaching essentially occurs via autotrophic and heterotrophic mechanisms pathways, with the former being either direct or indirect, whilst the latter being only purerly indirect [9][13], as depicted in Figure 1.
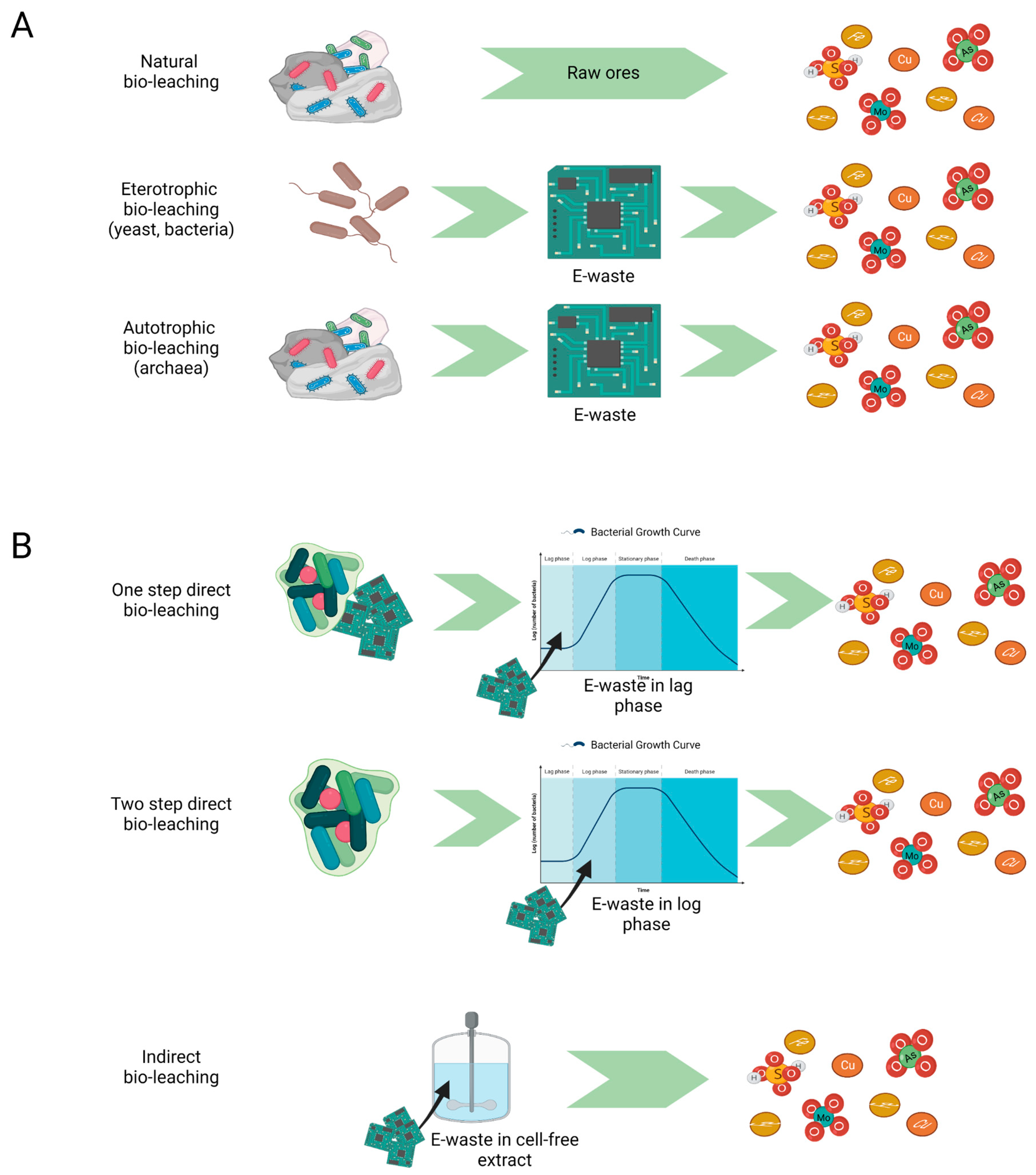
Figure 1. Bioleaching: mechanisms and strategies. (A) Heterotrophic and autotrophic bioleaching occurring in natural minerals deposits and in e-waste recovery processes resulting in free metal ions and metal oxides; (B) Direct (one-step and two-step) and indirect approach to bioleaching process. In direct bioleaching, e-waste is introduced during the initial microbial growth phase (one-step) or exponential growth phase (two-step). In indirect bioleaching, only microbial metabolites interact with e-waste.
Direct bioleaching implies the contact between microorganisms and metallic materials. This is valid for both single- and double-step processes, where the solid containing metals are mixed with microorganisms from the beginning or during the exponential microbial growth, respectively. Though direct and indirect bioleaching are both effective methods, the first one may run into the inhibition of microbial activity due to the toxicity of leached metals [13]. However, direct contact promotes the adhesion between microorganisms and the solid surface, mediated by the formation of biofilms and the uptake or complexation of metals through the excretion of bio-lixiviants and chemicals that perturb the environmental state, respectively, ending with the release of metal ions into solution. Recently, insights into the mechanism of interspecies communication within the archaeal biofilm and its regulation have enabled a deeper understanding of the bioleaching processes [15]. Autotrophic bioleaching uses carbon dioxide as a carbon source and iron Fe (II) or sulphur S (0) as the main energy sources to carry out their oxidation [16]. The general chemical reactions occurring in bioleaching are extensively reported in several studies [11][17][18] and more recently by Magoda K. and Mekuto L. (2022) [19]. Sulfur-oxidising bacteria (e.g., Acidithiobacillus thiooxidans,), iron- and sulphur-oxidising bacteria (Acidithiobacillus ferrooxidans) and iron-oxidising bacteria (Leptospirillum ferrooxidans) are the most employed autotrophs in bioleaching processes because of their resistance to heavy metal toxicity and their simple nutritional requirements [20]. .The consequent acidification due to the production of sulfuric acid and ferric ions causes metal dissolution, although for acid-insoluble sulphides or some metal oxides can be recovered either through biological oxidation, when the metal is soluble at a high oxidation state, or biological reduction, occurring when the metal is more soluble in a low oxidation state (e. g., Ni, V, and Mn) [21]. In contrast, heterotrophic bioleaching occurs via an indirect mechanism based on organic acids (e.g., citric or oxalic acid), ligands (e.g., chelators and siderophores) and exopolysaccharides (EPS) biosurfactants [15]. Among archaea, Haloferax mediterranei has been chosen as a model organism for EPS production, achieving a technological level of readiness (TRL) of 2 [22]. Heterotrophic bioleaching can be performed via acidolysis (Acidothiobacillus ferroxidans and Leptospirillum ferroxidans) or complexolysis (Chromobacterium violaceum, Pseudomonas aeruginosa, Pseudomonas flourescens, and Bascillus megaterium). In acidolysis, organic acids produce complexes with metal ions to enhance their mobilisation through a protonation mechanism, while complexolysis occurs when a chelating agent combines with metal on the surface or via biosurfactants acting as complexing agents. A third possible mechanism for heterotrophic bioleaching is the Fe (III) or (Mn (IV) dissimilatory reduction under anaerobic conditions in complex media, where the oxidation of organic matter is coupled to the metal reduction Shewenella putrfaciens is a reference microorganism for this process [23].
Bioleaching has been successfully applied in indirect and direct approaches for the recovering of valuable metals (i.e., Ni, Si, Cu, Ga, Mg, Te, Zn) from the dismissed light emitting diode (LED) [9], end-of-life PV solar cells [24], printed circuit board (PCB) [25] and other e-waste [1][26][27][28].
2. Interactions between Archaea and Metals
Microorganisms belonging to the Archaea domain are widely recognised as cosmopolitan ubiquitous organisms capable of adapting to either natural or anthropogenic environments characterised by extreme life conditions (i.e., natural gas and oil reservoirs, acid mines, hydrothermal vents) [29][30][31][32][33]. The relation between metals and thermophilic archaea stems from their natural ability to live at high temperatures and high-metals concentrations [7], thanks to unique cell wall structures, thermostable enzymes, and metabolic features [34].
Archaea also requires metals because of their role in enzymatic structures and co-factors, as final electron acceptors, or for sustaining their growth with metal enzymes and for pathways requiring metal ions as co-factor constituting the archaeal metallosome [31].
Genomic and proteomic studies allowed for the identification of similarities between archaeal, bacterial and eukaryotic metallosome, with the most common domains coding for Fe-, Co-, and Zn- binding proteins and only a low percentage coding for binding domains (<0.3%) for Ni-, Cu-, and Mo- [35]. Nonetheless, a higher percentage of Fe- binding domains were identified in archaeal genomes (≈7.1%) when compared to bacterial (≈3.9%) and eukaryotic (1.1%) ones. Although less represented, metal binding domains related to Ni-, Co- and Cu- utilisation/transport were also identified in archaeal genomes [35][36]. Table 1 resumes the roles that the previously mentioned metals have in Archaea.
Table 1. Physiological role of essential metals in some selected species of Archaea.
| Metal | Microorganism | Function in Archaea | References |
|---|---|---|---|
| Fe- | Halobacterium spp., Methanosarcina spp., Methanobacterium spp., Sulfolobus spp., Thermoplasma spp., Ferroplasma spp., Pyrobaculum spp. | Fe (II)oxidation, Fe (III) reduction, Fe4S4-ferredoxin, Fe4S4 cluster for S- adenosylmethionine cleavage, Ni-Fe hydrogenase | [31][35][37][38][39] |
| Zn- | n.s. | “Small proteins” class genes (Zn finger motifs and Really Interesting gene (RING)) | [35] |
| Co- | Methanosarcina spp., Sulfolobus solfataricus, Thermoplasma acidophilum | Found in co-enzyme B12 structure, Ni/Co uptake system | [40][41] |
| Ni- | Sulfolobus spp., Halobacter spp., Methanococcus spp. | Enzymatic co-factor for different enzymes:Ni-Fe hydrogenase, CO de-hydrogenase, methyl-CoM reductase, urease | [39][40][41] |
| Cu- | Halobacterium spp., Methanosarcina spp., Methanobacterium spp. | Copper-binding proteins, N2O reductase | [37] |
| Mo- | Sulfolobales spp., Halobacteriales spp., Methanosarcinales spp., Methanococcales spp., Methanomicrobiales spp. | Molybdenum co-factor (Moco) involved in W utilization | [42] |
With archaeal life requiring metals that are classified as CRMs (i.e., Co) and toxic materials (i.e., Ni, Co, Mo) and being capable of storing them in cellular structures (i.e., enzymes, co-factors, proteins) applications of archaeal cultures for metal detoxification and recovery surely sounds appealing. Furthermore, different studies have highlighted how archaeal-driven biotechnological applications (i.e., biomethanation, anaerobic digestion) need a stable supply of trace metals, including Fe, Co, Ni, Cu, and Mo, in order to maintain process stability [43][44][45][46][47].
A possible solution for economically viable metal recovery could be represented by its integration within already operating processes. In this logic, the modulation and fine-tuning of waste metals, supplied as nutrients, could sustain the systems requirements while increasing the sustainability of the process through the integration and valorisation of different waste streams.
Although most knowledge about the interaction of archaea with metals focuses on the role of Fe, Co, Ni, Cu, and Mo, many sources have reported interactions with non-essential metals, also listed as CRMs, precious and toxic elements (i.e., As, Cd, Pt, V, and U) [3][4][5][9]. A deeper insight regarding such archaea-metal interaction will be provided in further sections.
3. Bioleaching among Archaea
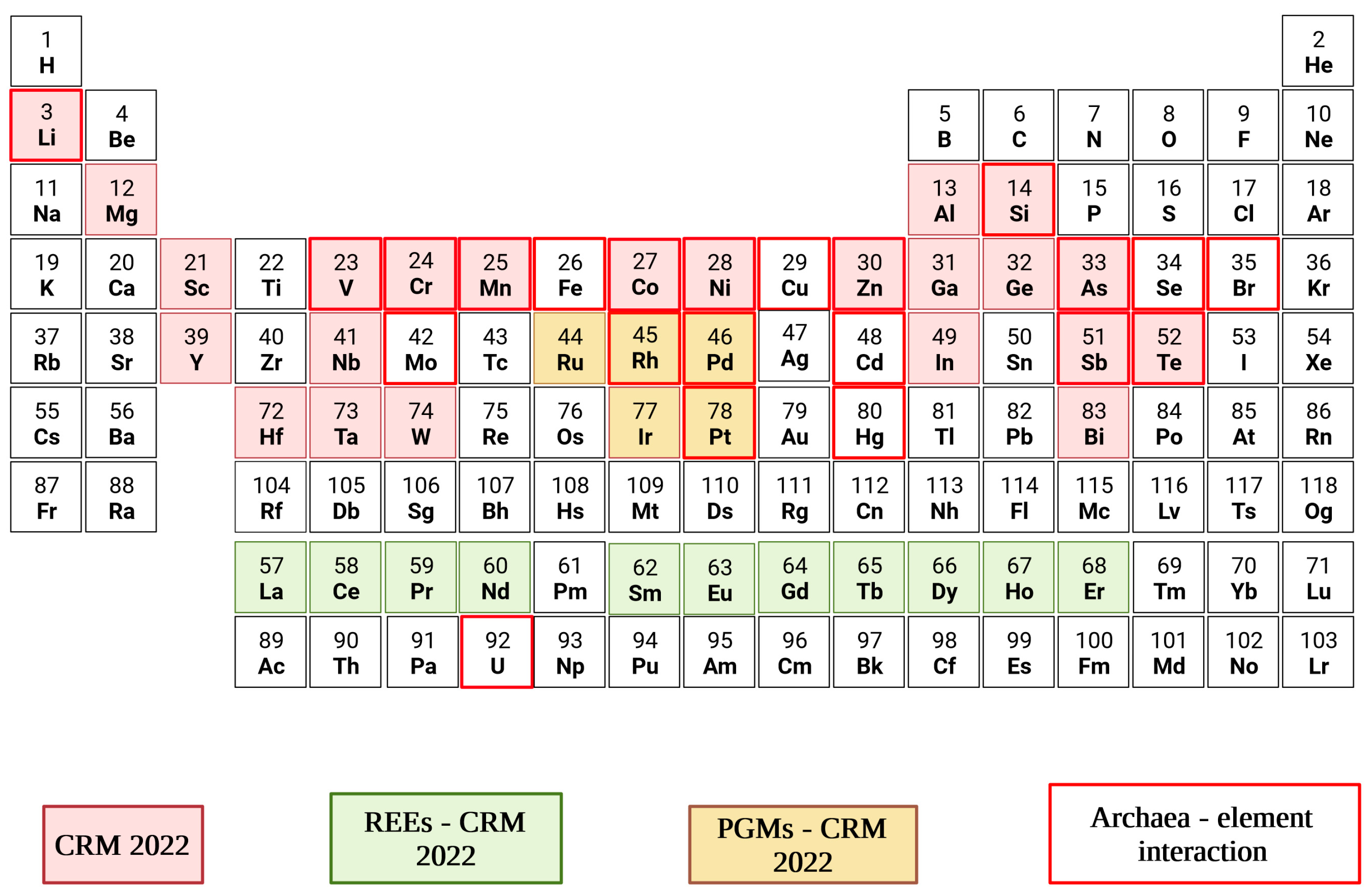
Despite the peculiar ubiquities and the great biotechnological potential of Archaea, Bacteria and Eukarya have always dominated the scene in terms of research studies, industrial application, and public perception. Despite Archaea's unusual ubiquity and tremendous biotechnological promise, Bacteria and Eukarya have traditionally dominated the scene in terms of study, industrial use, and popular perception. As detailed in the review authored by [22] on the current status of archaeal cell factories in bioproduction, few companies are investing in Archaea, while commercialisation is limited to extremophilic genera, which are suitable for applications in industrial processes under harsh conditions [48]. As regards the biomining and bioleaching industry, the engagement of Archaea is very limited, while research studies are mainly restricted to thermoacidophiles genera, often cultured with bacterial species whose leaching ability has already been validated [7][49]. However, archaea are also members of non-extreme environments [50]. Thus, the scenarios of possible industrial applications and their operating conditions are multivariate. A panoramic of CRMs typically contained in e-wastes and targeted by the Archaea herein discussed is presented in Figure 2.
3.1. Extremophile Archaeon
Several species of thermoacidophiles Archaea, such as Acidianus sp, Ferroplasma acidophilum, Metallosphaera sp, Metallosphaera sedula, have been identified in stirred-tank mineral bioleaching and bio-oxidation operations [53]. Other species isolated from a hydrothermal pool and mineral sulphide ores, respectively Acidiplasma aeolicum and Acidiplasma cupricumulans [54], can oxidise and reduce iron. The Sulfolobacaee family comprises species representative of acid hot spiring able to oxidise elemental sulphur (i.e., Sulfolobus acidocaldarius DSM 639, Sulfolobus solfataricus P2, Sulfolobus tokodaii JCM 10545, Stygiolobus azoricus DSM 6296) [55][56]. Among Thermoplasmatales, researchers can mention Picrophilus torridus and Thermogymnomonas acidicola JCM 13583, which inhabit dry solfataric fields [57][58], Thermoplasma acidophilum DSM 1728 and Thermoplasma volcanium GSS1 [59], respectively identified from coal refuse piles and hot acid springs. Novel species isolated from acid mine drainage (AMD), such as Cuniculiplasma divulgatum JCM 30642, Candidatus Micrarchaeum acidiphilum ARMAN-2 NIA Candidatus Parvarchaeum acidiphilum ARMAN-4 NIA Candidatus Parvarchaeum acidiphilum ARMAN-5 are likely capable of oxidising metal sulphides [12].
Heavy metals, such as As, Hg, and Cd, can be remediated by several extremophilic archaeon species [21]. Sulfolobus acidocaldarius strain BC was isolated in an acidic, sulfuric thermal spring in the Yellowstone National Park and recognised to be capable of oxidising arsenite As (IV) to arsenate As (V) [60]. Other Archaea strains, such as Aeropyrum pernix K1, Pyrobaculum calidifontis JCM 11548, and Sulfolobus tokodaii 7 and the genus Halorubrum contain arsenite oxidase genes [61][62][63]. Sulfolobus solfataricus species, Halococcus, Halobacterium, and, to a lesser extent, Haloferax genera were capable of mercury Hg (II) volatilisation into Hg (0), thanks to the presence of mercury reductase genes [64][65][66]. Halobacterium noricense stands out for its ability to adsorb cadmium [67], while Haloferax strain BBK2 uptakes cadmium intracellularly [68]. Among metalloids included in the CRMs list, Si is of primary importance given its large application in PV solar cells Si is crucial due to its widespread use in PV solar cells.. The hyperthermophilic sulphur-metabolising Pyrococcus abyssi was fossilised during and after exposure to a silica-saturated solution (about 500 ppm of SiO2) in a simulated hydrothermal environment [69], demonstrating the ability to bind silica at the S-layer sites and integrate it in replacement of the cell wall.
Based on the above-mentioned studies, thermophilic and hyperthermophilic archaea can be applied in many types of high-temperature metal-contaminated waste streams, while haloarchaea can be used in the treatment of hypersaline environments and wastewaters for heavy metal removal.
3.2. Methanogenic Archaea
Methanogens, which already play an important role in the global methane (CH4) cycle by producing CH4 via methanogenesis, can also recover many critical, platinum group metal (PGM) and non-critical metals from the —end-life products, mining residues and waste streams.
Consortia
It has been stated in many studies that anaerobic granular sludge is efficient in PGM metals recovery (Figure 2). The work by Espadas et al. reports Pd (II) removal via reduction to Pd (0) nanoparticles or biosorption into the biomass of a methanogenic granular sludge from a full-scale up-flow anaerobic sludge blanket (UASB) reactor processing brewery wastewater [70]. While the reduction route was supported using external H2, formate, and ethanol, the biosorption dominated when acetate, lactate and pyruvate were added as external electron donors. A more recent study determined the ability to reduce Pt (II) and Pt (IV) to Pt (0) with the same anaerobic sludge applied by Pat-Espadas et al. 2018 [70]. Rh (III), which is one of the rarest elements in the geosphere (about 0.001 ppm), was also reduced to Rh (0) nanoparticles, with ethanol as an external donor of electrons [71]. Ramos-Ruiz et al., 2016 highlighted the potential of anaerobic granular sludge from a methanogenic reactor as a biocatalyst for the reduction of both Te (IV) and Te (VI) to produce Te (0) nanoparticles [72]. Methanobacterium genus was detected as predominant among the archaeal community populating a fluidised bed reactor applied to lithium-ion batteries (LIBs) leaching via biogenic hydrogen sulphide [73].
Pure Culture in H2/CO2
Compared to consortia, pure culture applications present some advantages in terms of process reproducibility and simplicity. However, while pure cultures usually require more strict conditions (e.g., nutrient demand and key process parameters), consortia are typically more robust [43]. The mesophilic hydrogenotrophic strain Methanobacterium bryatii BKYH has been isolated from a copper mining area in the Upper Peninsula of Michigan and is able to chelate Cu by secreting a specific protein in response to copper exposure [74]. Increased Cu concentration in the medium results in the production of an extracellular Cu response- (CRX) protein encoded by the crx gene and promoted by an archaeal Cu-responsive promoter, which could be synthesised as part of a more generic stress response [75].
Zhang et al., 2014 published the first study reporting methanogenic archaeal strains as capable of reducing vanadium (V) [76], classified as CRM in 2022 (Figure 2). Methanosarcina mazei and Methanothermobacter thermautotrophicus could reduce up to 10 mM and 5 mM of vanadate V(V) to vanadyl V(IV), respectively, inducing solid extracellular precipitation as the bioreduction occurred at the cell membrane level. However, at some point, methanogenesis stops with vanadyl generation, possibly due to the redirection of electrons from methanogenesis to vanadate reduction.
According to Singh et al., 2016 Methanothermobacter thermautotrophicus is capable of reducing Co (III) present at a maximum concentration of 4 mM [77] and can reduce up to 1 mM of Cr (VI) to Cr (III) without an inhibitory effect on methanogenesis and cell growth through an intracellular and extracellular reduction mechanism [78]. Holmes et al. reported the potential role of acetoclastic methanogens belonging to the genus Methanosarcina in reducing U(VI) present in contaminated groundwater [79]. After a period of acetate amendment in the field, the predominance of Methanosarcina corresponded to methane accumulation and U(VI) reduction. Like Pyrococcus abyssi, the hyperthermophilic, hydrogenotrophic methanogen Methanocaldococcus jannaschii could bind Si on its cell wall and accomplish the silicification mechanism, but only when metal cations, particularly Fe (III), were present [80]. However, a more detailed comprehension of this mechanism is pivotal in order to understand its exploitability in the recovery of Si-rich wastes.
3.3. Adverse Interactions between Methanogens and Critical Metals
Despite both methanogenic consortia and pure cultures have been proven capable of reducing different metals, studies have also reported potential issues related to the metal-methanogen interactions as:
-
Inhibition and toxicity: 50% inhibiting concentrations (IC50) for Cd, selenite, tellurite, and tellurate have been investigated on methanogenic consortia, with acetoclastic methanogens displaying higher IC50 (IC50Cd 8.6 mg/L, IC50selenite 24.1 mg/L, IC50tellurite 8.6 mg/L, IC50tellurate 10.2 mg/L) than those of hydrogenotrophic methanogens (IC50Cd 2.9 mg/L, IC50selenite 18 mg/L, IC50tellurite 8.6 mg/L, IC50tellurate 10.2 mg/L) [81]. Similarly, IC50 for Pd(II), Pt(II) and Pt(IV) for methanogens were reported at 2.7, 2.4 and 3.7 mg/L, respectively [70], whilst full inhibition of methanogenesis occurred at Pt(II) concentration higher than 5 mg/L [82];
-
Competition between metal reduction and methanogenesis: Methanogenesis, both in anaerobic digestion and in the biomethanation process, requires electron donors to reduce CO2. When considering the bioleaching process, the addition of an external electron donor is often needed due to the competition for electrons that are redirected from methanogenesis to metal reduction [76]. In an anaerobic granular sludge, endogenous substrates can provide sufficient electron equivalents for the leaching of metals of interest, although reduction rates may increase with excess electrons, as reported by [72]. Among external electron donors to be added to methanogenic anaerobic sludge, ethanol is considered a safe and economical option. Fermentation of ethanol by the acetogenic community populating the granular sludge generates H2 that is then redirected to metal reduction. However, it should be mentioned that some studies have described the abiotic reduction of some metals through direct chemical reduction by H2 and formate [70][82][83].
3.4. Anaerobic Methanotrophic Archaea (ANME)
Microorganisms play an important role in the global CH4 cycle that is controlled by the balance between anaerobic production via methanogenesis and CH4 removal via methanotrophic oxidation. Methane oxidation is anaerobic (AOM) in more than 90% of cases [84]. AOM can be coupled to other electron acceptors besides the well-known sulphates SO42−, such as nitrates NO3− (N-DAMO), or metals such as manganese Mn (IV), iron Fe (III), As (V), Cr (VI), Se (VI), Sb (V), V (V), and Br (V) [85] through the establishment of a syntrophic partnership as electron sinks. However, it is not fully elucidated whether the reduction of metallic electron acceptors is independent or supported by electron transfer to syntrophic partners, interspecies electron transfer, nanowires or conductive pili [86].
The study by Zhang et al., 2020 investigated the bio-reduction of vanadate V(V) present in groundwater using an anaerobic sludge inoculum taken from a wastewater treatment resource [87]. Biological mediated vanadate removal, corresponding to 95.8 ± 3.1% of 1 mM, occurred after seven days of incubation using CH4 as the sole electron donor. Microbial community analysis revealed a more significative change in the archaeal population than in the inoculum, with a massive presence of the Methanobacterium genus, which is known to oxidise methane via reverse methanogenesis, and the hydrogenotrophic methanogens belonging to Methanomassiliicoccus. Thus, vanadate reduction occurred through the anaerobic oxidation of methane and synergistic interactions with methane-oxidising bacteria such as Methylomonas. Evidence of the reduction of antimonate Sb (V) to Sb (III) using CH4 is reported by the work of Lai et al., 2018 [85] in a membrane biofilm batch reactor inoculated with a pre-enriched culture with methane and Sb (V), Figure 2. Increasing concentrations of Sb (V), 0.41, 0.82, 1.6 mM, at different stages of the experiment were 100% biologically reduced in the form of Sb2O3 crystal precipitates. The Archaea community was widely enriched during the different operational stages, where Methanosarcina and Methanolobus genera were the two most abundant methanogens, with the former being predominant. Other studies suggested a key role of the methanogenic archaea Methanosarcina in AOM via reverse methanogenesis, as in Luo et al., 2017 where the electron acceptor was Br (V) in the form of BrO3− [88]. A possible explanation is the close phylogenetic relationship between ANME-2a/b, ANME-2c, ANME-2d, and ANME-3 and Methanosarcinales [89]. The archaea family ANME-2d, also knowns as Methanoperedenaceae lineage, was found to be able to oxidise methane through Mn (V) reduction in a bioreactor supplied with CH4 and pulse of birnessite operating for 480 days [90]. The inoculum originated from a bioreactor inoculated with freshwater sediment and performing AOM via Fe (III) reduction. The average Mn (II) production rate was 0.185 mmol L−1 day−1 with a methane consumption rate of 0.045 mmol L−1 day−1.
References
- Mishra, S.; Panda, S.; Akcil, A.; Dembele, S.; Agcasulu, I. A Review on Chemical versus Microbial Leaching of Electronic Wastes with Emphasis on Base Metals Dissolution. Minerals 2021, 11, 1255.
- Chen, Z.; Liu, L.; Wang, H.; Liu, L.; Wang, X. Pyrolysis Characteristics and Non-Isothermal Kinetics of Integrated Circuits. Materials 2022, 15, 4460.
- Mirazimi, S.; Abbasalipour, Z.; Rashchi, F. Vanadium removal from LD converter slag using bacteria and fungi. J. Environ. Manag. 2015, 153, 144–151.
- Zhang, B.; Li, Y.; Fei, Y.; Cheng, Y. Novel Pathway for Vanadium(V) Bio-Detoxification by Gram-Positive Lactococcus raffinolactis. Environ. Sci. Technol. 2021, 55, 2121–2131.
- Willner, J.; Fornalczyk, A.; Gajda, B.; Saternus, M. Bioleaching of indium and tin from used LCD panels. Physicochem. Probl. Miner. Process. 2018, 54, 639–645.
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Molecular, Clinical and Environmental Toxicology: Volume 3: Environmental Toxicology; Springer Science & Business Media: New York, NY, USA, 2012; Volume 101, pp. 1–30.
- Ranawat, P.; Rawat, S. Metal-tolerant thermophiles: Metals as electron donors and acceptors, toxicity, tolerance and industrial applications. Environ. Sci. Pollut. Res. 2017, 25, 4105–4133.
- Valls, M.; De Lorenzo, V. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol. Rev. 2002, 26, 327–338.
- Pourhossein, F.; Mousavi, S.M. A novel step-wise indirect bioleaching using biogenic ferric agent for enhancement recovery of valuable metals from waste light emitting diode (WLED). J. Hazard. Mater. 2019, 378, 120648.
- Johnson, D.B. Development and application of biotechnologies in the metal mining industry. Environ. Sci. Pollut. Res. 2013, 20, 7768–7776.
- Baniasadi, M.; Vakilchap, F.; Bahaloo-Horeh, N.; Mousavi, S.M.; Farnaud, S. Advances in bioleaching as a sustainable method for metal recovery from e-waste: A review. J. Ind. Eng. Chem. 2019, 76, 75–90.
- Castro, C.; Urbieta, M.; Cazón, J.P.; Donati, E. Metal biorecovery and bioremediation: Whether or not thermophilic are better than mesophilic microorganisms. Bioresour. Technol. 2019, 279, 317–326.
- Sarkodie, E.K.; Jiang, L.; Li, K.; Yang, J.; Guo, Z.; Shi, J.; Deng, Y.; Liu, H.; Jiang, H.; Liang, Y.; et al. A review on the bioleaching of toxic metal(loid)s from contaminated soil: Insight into the mechanism of action and the role of influencing factors. Front. Microbiol. 2022, 13.
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–643.
- van Wolferen, M.; Orell, A.; Albers, S.-V. Archaeal biofilm formation. Nat. Rev. Genet. 2018, 16, 699–713.
- Yu, Z.; Han, H.; Feng, P.; Zhao, S.; Zhou, T.; Kakade, A.; Kulshrestha, S.; Majeed, S.; Li, X. Recent advances in the recovery of metals from waste through biological processes. Bioresour. Technol. 2020, 297, 122416.
- Bosecker, K. Bioleaching: Metal solubilization by microorganisms. FEMS Microbiol. Rev. 1997, 20, 591–604.
- Faramarzi, M.A.; Mogharabi-Manzari, M.; Brandl, H. Bioleaching of metals from wastes and low-grade sources by HCN-forming microorganisms. Hydrometallurgy 2019, 191, 105228.
- Magoda, K.; Mekuto, L. Biohydrometallurgical Recovery of Metals from Waste Electronic Equipment: Current Status and Pro-posed Process. Recycling 2022, 7, 67.
- Roy, J.J.; Cao, B.; Madhavi, S. A review on the recycling of spent lithium-ion batteries (LIBs) by the bioleaching approach. Chemosphere 2021, 282, 130944.
- Krzmarzick, M.J.; Taylor, D.K.; Fu, X.; McCutchan, A.L. Diversity and niche of archaea in bioremediation. Hindawi 2018, 2018, 3194108.
- Pfeifer, K.; Ergal, I.; Koller, M.; Basen, M.; Schuster, B.; Rittmann, S.K.-M. Archaea Biotechnology. Biotechnol. Adv. 2020, 47, 107668.
- Lovley, D.R.; Holmes, D.E.; Nevin, K.P. Dissimilatory Fe(III) and Mn(IV) Reduction. Adv. Microb. Physiol. 2004, 49, 219–286.
- Chakankar, M.; Su, C.H.; Hocheng, H. Leaching of metals from end-of-life solar cells. Environ. Sci. Pollut. Res. 2018, 26, 29524–29531.
- Priya, A.; Hait, S. Extraction of Metals from High Grade Waste Printed Circuit Board by Conventional and Hybrid Bioleaching Using Acidithiobacillus Ferrooxidans. Hydrometallurgy 2018, 177, 132–139.
- Priya, A.; Hait, S. Feasibility of Bioleaching of Selected Metals from Electronic Waste by Acidiphilium acidophilum. Waste Biomass- Valorization 2017, 9, 871–877.
- Priya, A.; Hait, S. Comparative assessment of metallurgical recovery of metals from electronic waste with special emphasis on bioleaching. Environ. Sci. Pollut. Res. 2017, 24, 6989–7008.
- Sinha, R.; Chauhan, G.; Singh, A.; Kumar, A.; Acharya, S. A novel eco-friendly hybrid approach for recovery and reuse of copper from electronic waste. J. Environ. Chem. Eng. 2018, 6, 1053–1061.
- Liu, Y.; Whitman, W.B. Metabolic, Phylogenetic, and Ecological Diversity of the Methanogenic Archaea. Ann. New York Acad. Sci. 2008, 1125, 171–189.
- Basso, O.; Lascourreges, J.-F.; Le Borgne, F.; Le Goff, C.; Magot, M. Characterization by culture and molecular analysis of the microbial diversity of a deep subsurface gas storage aquifer. Res. Microbiol. 2009, 160, 107–116.
- Bini, E. Archaeal transformation of metals in the environment. FEMS Microbiol. Ecol. 2010, 73, 1–16.
- Oren, A. Taxonomy of halophilic Archaea: Current status and future challenges. Extremophiles 2014, 18, 825–834.
- Baker, B.J.; De Anda, V.; Seitz, K.W.; Dombrowski, N.; Santoro, A.E.; Lloyd, K.G. Diversity, ecology and evolution of Archaea. Nat. Microbiol. 2020, 5, 887–900.
- Sar, P.; Kazy, S.K.; Paul, D.; Sarkar, A. Metal Bioremediation by Thermophilic Microorganisms. In Thermophilic Microbes in Environmental and Industrial Biotechnology: Biotechnology of Thermophiles; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 9789400758995.
- Dupont, C.L.; Yang, S.; Palenik, B.; Bourne, P.E. Modern proteomes contain putative imprints of ancient shifts in trace metal geochemistry. Proc. Natl. Acad. Sci. USA 2006, 103, 17822–17827.
- Andreini, C.; Banci, L.; Bertini, I.; Elmi, S.; Rosato, A. Non-Heme Iron Through the Three Domains of Life. Proteins: Struct. Funct. Bioinform. 2007, 67, 317–324.
- Andreini, C.; Banci, L.; Bertini, I.; Rosato, A. Occurrence of Copper Proteins through the Three Domains of Life: A Bioinformatic Approach. J. Proteome Res. 2007, 7, 209–216.
- Lyu, Z.; Chou, C.-W.; Shi, H.; Wang, L.; Ghebreab, R.; Phillips, D.; Yan, Y.; Duin, E.C.; Whitman, W.B. Assembly of Methyl Coenzyme M Reductase in the Methanogenic Archaeon Methanococcus maripaludis. J. Bacteriol. 2018, 200.
- Tsurumaru, H.; Ito, N.; Mori, K.; Wakai, S.; Uchiyama, T.; Iino, T.; Hosoyama, A.; Ataku, H.; Nishijima, K.; Mise, M.; et al. An extracellular hydrogenase mediating iron corrosion is encoded in a genetically unstable genomic island in Methanococcus maripaludis. Sci. Rep. 2018, 8, 15149.
- Rodionov, D.A.; Hebbeln, P.; Gelfand, M.; Eitinger, T. Comparative and Functional Genomic Analysis of Prokaryotic Nickel and Cobalt Uptake Transporters: Evidence for a Novel Group of ATP-Binding Cassette Transporters. J. Bacteriol. 2006, 188, 317–327.
- Zhang, Y.; A Rodionov, D.; Gelfand, M.S.; Gladyshev, V.N. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genom. 2009, 10, 78.
- Zhang, Y.; Gladyshev, V.N. Molybdoproteomes and Evolution of Molybdenum Utilization. J. Mol. Biol. 2008, 379, 881–899.
- Bellini, R.; Bassani, I.; Vizzarro, A.; Azim, A.A.; Vasile, N.S.; Pirri, C.F.; Verga, F.; Menin, B. Biological Aspects, Advancements and Techno-Economical Evaluation of Biological Methanation for the Recycling and Valorization of CO2. Energies 2022, 15, 4064.
- Romero-Güiza, M.; Vila, J.; Mata-Alvarez, J.; Chimenos, J.; Astals, S. The role of additives on anaerobic digestion: A review. Renew. Sustain. Energy Rev. 2016, 58, 1486–1499.
- Schattauer, A.; Abdoun, E.; Weiland, P.; Plöchl, M.; Heiermann, M. Abundance of trace elements in demonstration biogas plants. Biosyst. Eng. 2010, 108, 57–65.
- Wintsche, B.; Jehmlich, N.; Popp, D.; Harms, H.; Kleinsteuber, S. Metabolic Adaptation of Methanogens in Anaerobic Digesters Upon Trace Element Limitation. Front. Microbiol. 2018, 9, 405.
- Wintsche, B.; Glaser, K.; Sträuber, H.; Centler, F.; Liebetrau, J.; Harms, H.; Kleinsteuber, S. Trace Elements Induce Predominance among Methanogenic Activity in Anaerobic Digestion. Front. Microbiol. 2016, 7, 2034.
- Schiraldi, C.; Giuliano, M.; De Rosa, M. Perspectives on biotechnological applications of archaea. Archaea 2002, 1, 75–86.
- Straub, C.T.; A Counts, J.; Nguyen, D.M.N.; Wu, C.-H.; Zeldes, B.M.; Crosby, J.R.; Conway, J.M.; Otten, J.K.; Lipscomb, G.L.; Schut, G.J.; et al. Biotechnology of extremely thermophilic archaea. FEMS Microbiol. Rev. 2018, 42, 543–578.
- Hedlund, B.P.; Zhang, C.; Wang, F.; Rinke, C.; Martin, W.F. Editorial: Ecology, Metabolism and Evolution of Archaea-Perspectives from Proceedings of the International Workshop on Geo-Omics of Archaea. Front. Microbiol. 2022, 12.
- Energy Act of 2020 (Critical Minerals Provisions). Available online: https://www.iea.org/policies/16065-energy-act-of-2020-critical-minerals-provisions (accessed on 2 February 2023).
- Direzione generale del Mercato interno, dell’industria, dell’imprenditoria e delle P (Commissione Europea). Blengini, G.A., el Latunussa, C., Eynard, U., Torres De Matos, C., Wittmer, D., Georgitzikis, K., Pavel, C., Carrara, S., Mancini, L., et al., Eds.; Study on the EU’s List of Critical Raw Materials (2020); Final Report; Commissione Europea: Brussels, Belgium, 2020.
- Siezen, R.J.; Wilson, G. Bioleaching genomics. Microb. Biotechnol. 2009, 2, 297–303.
- Golyshina, O.V.; Yakimov, M.M.; Lünsdorf, H.; Ferrer, M.; Nimtz, M.; Timmis, K.N.; Wray, V.; Tindall, B.J.; Golyshin, P.N. Acidiplasma aeolicum gen. nov., sp. nov., a euryarchaeon of the family Ferroplasmaceae isolated from a hydrothermal pool, and transfer of Ferroplasma cupricumulans to Acidiplasma cupricumulans comb. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 2815–2823.
- Brock, T.D.; Brock, K.M.; Belly, R.T.; Weiss, R.L. Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Microbiol. 1972, 84, 54–68.
- Segerer, A.H.; Trincone, A.; Gahrtz, M.; Stetter, K.O. Stygiolobus azoricus gen. nov., sp. nov. Represents a Novel Genus of Anaerobic, Extremely Thermoacidophilic Archaebacteria of the Order Sulfolobales. Int. J. Syst. Evol. Microbiol. 1991, 41, 495–501.
- Fütterer, O.; Angelov, A.; Liesegang, H.; Gottschalk, G.; Schleper, C.; Schepers, B.; Dock, C.; Antranikian, G.; Liebl, W. Genome sequence of Picrophilus torridus and its implications for life around pH 0. Proc. Natl. Acad. Sci. 2004, 101, 9091–9096.
- Itoh, T.; Yoshikawa, N.; Takashina, T. Thermogymnomonas acidicola gen. nov., sp. nov., a novel thermoacidophilic, cell wall-less archaeon in the order Thermoplasmatales, isolated from a solfataric soil in Hakone, Japan. Int. J. Syst. Evol. Microbiol. 2007, 57, 2557–2561.
- Segerer, A.; Langworthy, T.A.; Stetter, K.O. Thermoplasma acidophilum and Thermoplasma volcanium sp. nov. from Solfatara Fields. Syst. Appl. Microbiol. 1988, 10, 161–171.
- Sehlin, H.M.; Lindstrã¶m, E.B. Oxidation and reduction of arsenic by Sulfolobus acidocaldarius strain BC. FEMS Microbiol. Lett. 1992, 93, 87–92.
- Lebrun, E.; Brugna, M.; Baymann, F.; Muller, D.; Lièvremont, D.; Lett, M.-C.; Nitschke, W. Arsenite Oxidase, an Ancient Bioenergetic Enzyme. Mol. Biol. Evol. 2003, 20, 686–693.
- Heinrich-Salmeron, A.; Cordi, A.; Brochier-Armanet, C.; Halter, D.; Pagnout, C.; Abbaszadeh-Fard, E.; Montaut, D.; Seby, F.; Bertin, P.N.; Bauda, P.; et al. Unsuspected Diversity of Arsenite-Oxidizing Bacteria as Revealed by Widespread Distribution of the aoxB Gene in Prokaryotes. Appl. Environ. Microbiol. 2011, 77, 4685–4692.
- Ordoñez, O.F.; Rasuk, M.C.; Soria, M.N.; Contreras, M.; Farías, M.E. Haloarchaea from the Andean Puna: Biological Role in the Energy Metabolism of Arsenic. Microb. Ecol. 2018, 76, 695–705.
- Wang, Y.; Boyd, E.; Crane, S.; Lu-Irving, P.; Krabbenhoft, D.; King, S.; Dighton, J.; Geesey, G.; Barkay, T. Environmental Conditions Constrain the Distribution and Diversity of Archaeal merA in Yellowstone National Park, Wyoming, U.S.A. Microb. Ecol. 2011, 62, 739–752.
- Schelert, J.; Dixit, V.; Hoang, V.; Simbahan, J.; Drozda, M.; Blum, P. Occurrence and Characterization of Mercury Resistance in the Hyperthermophilic Archaeon Sulfolobus solfataricus by Use of Gene Disruption. J. Bacteriol. 2004, 186, 427–437.
- Al-Mailem, D.M.; Al-Awadhi, H.; Sorkhoh, N.A.; Eliyas, M.; Radwan, S.S. Mercury resistance and volatilization by oil utilizing haloarchaea under hypersaline conditions. Extremophiles 2010, 15, 39–44.
- Showalter, A.R.; Szymanowski, J.E.S.; Fein, J.B.; A Bunker, B. An x-ray absorption spectroscopy study of Cd binding onto a halophilic archaeon. J. Phys. Conf. Ser. 2016, 712, 12079.
- Das, D.; Salgaonkar, B.B.; Mani, K.; Braganca, J.M. Cadmium resistance in extremely halophilic archaeon Haloferax strain BBK2. Chemosphere 2014, 112, 385–392.
- Orange, F.; Westall, F.; Disnar, J.-R.; Prieur, D.; Bienvenu, N.; LE Romancer, M.; Défarge, C. Experimental silicification of the extremophilic Archaea Pyrococcus abyssi and Methanocaldococcus jannaschii: Applications in the search for evidence of life in early Earth and extraterrestrial rocks. Geobiology 2009, 7, 403–418.
- Pat-Espadas, A.M.; Field, J.A.; Otero-Gonzalez, L.; Razo-Flores, E.; Cervantes, F.J.; Sierra-Alvarez, R. Recovery of palladium(II) by methanogenic granular sludge. Chemosphere 2016, 144, 745–753.
- Zhu, K. Chemical and Microbial Processes for Rhodium Recovery in the Graduate College. Ph.D. Thesis, The University of Arizona, Tucson, Arizona, 2022.
- Ramos-Ruiz, A.; Field, J.A.; Wilkening, J.V.; Sierra-Alvarez, R. Recovery of Elemental Tellurium Nanoparticles by the Reduction of Tellurium Oxyanions in a Methanogenic Microbial Consortium. Environ. Sci. Technol. 2016, 50, 1492–1500.
- Calvert, G.; Kaksonen, A.H.; Cheng, K.Y.; Van Yken, J.; Chang, B.; Boxall, N.J. Recovery of Metals from Waste Lithium Ion Battery Leachates Using Biogenic Hydrogen Sulfide. Minerals 2019, 9, 563.
- Kim, B.K.; Pihl, T.D.; Reeve, J.N.; Daniels, L. Purification of the copper response extracellular proteins secreted by the copper-resistant methanogen Methanobacterium bryantii BKYH and cloning, sequencing, and transcription of the gene encoding these proteins. J. Bacteriol. 1995, 177, 7178–7185.
- Macario, A.J.L.; Lange, M.; Ahring, B.K.; De Macario, E.C. Stress Genes and Proteins in the Archaea. Microbiol. Mol. Biol. Rev. 1999, 63, 923–967.
- Zhang, J.; Dong, H.; Zhao, L.; McCarrick, R.; Agrawal, A. Microbial reduction and precipitation of vanadium by mesophilic and thermophilic methanogens. Chem. Geol. 2014, 370, 29–39.
- Singh, R.; Dong, H.; Liu, D.; Marts, A.R.; Tierney, D.L.; Almquist, C.B. −reduction by thermophilic methanogen Methanothermobacter thermautotrophicus. Chem. Geol. 2015, 411, 49–56.
- Singh, R.; Dong, H.; Liu, D.; Zhao, L.; Marts, A.R.; Farquhar, E.; Tierney, D.L.; Almquist, C.B.; Briggs, B.R. Reduction of Hexavalent Chromium by the Thermophilic Methanogen Methanothermobacter thermautotrophicus. Geochim. Et Cosmochim. Acta 2015, 148, 442–456.
- E Holmes, D.; Orelana, R.; Giloteaux, L.; Wang, L.-Y.; Shrestha, P.; Williams, K.; Lovley, D.R.; Rotaru, A.-E. Potential for Methanosarcina to Contribute to Uranium Reduction during Acetate-Promoted Groundwater Bioremediation. Microb. Ecol. 2018, 76, 660–667.
- Orange, F.; Disnar, J.-R.; Westall, F.; Prieur, D.; Baillif, P. Metal cation binding by the hyperthermophilic microorganism, Archaea Methanocaldococcus Jannaschii, and its effects on silicification. Palaeontology 2011, 54, 953–964.
- Ramos-Ruiz, A.; Zeng, C.; Sierra-Alvarez, R.; Teixeira, L.H.; Field, J.A. Microbial Toxicity of Ionic Species Leached from the II-VI Semiconductor Materials, Cadmium Telluride (CdTe) and Cadmium Selenide (CdSe). Chemosphere 2016, 162, 131–138.
- Simon-Pascual, A.; Sierra-Alvarez, R.; Field, J.A. Platinum(II) reduction to platinum nanoparticles in anaerobic sludge. J. Chem. Technol. Biotechnol. 2018, 94, 468–474.
- Rotaru, A.-E.; Jiang, W.; Finster, K.; Skrydstrup, T.; Meyer, R.L. Non-enzymatic palladium recovery on microbial and synthetic surfaces. Biotechnol. Bioeng. 2012, 109, 1889–1897.
- Knittel, K.; Boetius, A.; Offre, P.; Spang, A.; Schleper, C.; Strous, M.; Jetten, M.S.; Pearson, A.; Ingalls, A.E.; Boucher, Y.; et al. Anaerobic Oxidation of Methane: Progress with an Unknown Process. Annu. Rev. Microbiol. 2009, 63, 311–334.
- Lai, C.-Y.; Dong, Q.-Y.; Rittmann, B.E.; Zhao, H.-P. Bioreduction of Antimonate by Anaerobic Methane Oxidation in a Membrane Biofilm Batch Reactor. Environ. Sci. Technol. 2018, 52, 8693–8700.
- Goyal, N.; Zhou, Z.; Karimi, I.A. Metabolic processes of Methanococcus maripaludis and potential applications. Microb. Cell Factories 2016, 15, 1–19.
- Zhang, B.; Jiang, Y.; Zuo, K.; He, C.; Dai, Y.; Ren, Z.J. Microbial vanadate and nitrate reductions coupled with anaerobic methane oxidation in groundwater. J. Hazard. Mater. 2019, 382, 121228.
- Luo, J.-H.; Wu, M.; Yuan, Z.; Guo, J. Biological Bromate Reduction Driven by Methane in a Membrane Biofilm Reactor. Environ. Sci. Technol. Lett. 2017, 4, 562–566.
- Wang, Y.; Xie, R.; Hou, J.; Lv, Z.; Li, L.; Hu, Y.; Huang, H.; Wang, F. The late Archaean to early Proterozoic origin and evolution of anaerobic methane-oxidizing archaea. Mlife 2022, 1, 96–100.
- Leu, A.O.; Cai, C.; McIlroy, S.J.; Southam, G.; Orphan, V.J.; Yuan, Z.; Hu, S.; Tyson, G.W. Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae. ISME J. 2020, 14, 1030–1041.
More
Information
Subjects:
Biotechnology & Applied Microbiology; Green & Sustainable Science & Technology; Metallurgy & Metallurgical Engineering
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
790
Revisions:
4 times
(View History)
Update Date:
04 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




