Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Bhupendra Koul | -- | 3306 | 2023-04-27 10:39:18 | | | |
| 2 | Camila Xu | + 1 word(s) | 3307 | 2023-04-28 03:52:45 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Khan, A.; Nasim, N.; Pudhuvai, B.; Koul, B.; Upadhyay, S.K.; Sethi, L.; Dey, N. Plant Synthetic Promoters Structure and Function. Encyclopedia. Available online: https://encyclopedia.pub/entry/43557 (accessed on 08 February 2026).
Khan A, Nasim N, Pudhuvai B, Koul B, Upadhyay SK, Sethi L, et al. Plant Synthetic Promoters Structure and Function. Encyclopedia. Available at: https://encyclopedia.pub/entry/43557. Accessed February 08, 2026.
Khan, Ahamed, Noohi Nasim, Baveesh Pudhuvai, Bhupendra Koul, Santosh Kumar Upadhyay, Lini Sethi, Nrisingha Dey. "Plant Synthetic Promoters Structure and Function" Encyclopedia, https://encyclopedia.pub/entry/43557 (accessed February 08, 2026).
Khan, A., Nasim, N., Pudhuvai, B., Koul, B., Upadhyay, S.K., Sethi, L., & Dey, N. (2023, April 27). Plant Synthetic Promoters Structure and Function. In Encyclopedia. https://encyclopedia.pub/entry/43557
Khan, Ahamed, et al. "Plant Synthetic Promoters Structure and Function." Encyclopedia. Web. 27 April, 2023.
Copy Citation
A promoter is the region of genomic DNA located upstream of a gene that initiates the process of transcription under specific cellular conditions. Structurally, it is modular in nature and comprises a core promoter that includes a TATA box and a CAAT box, as well as proximal and distal regions.
synthetic promoter
CRISPR
Cis-engineering
transcription factors
1. Introduction
Genetic engineering has emerged as a promising molecular discipline that can efficiently resolve the existing agricultural problems related to the escalating global population and can boost the agricultural productivity. Assuming that the success of transgenics depends on the expression level of the transgene, researchers in the field of plant synthetic promoter engineering are striving towards the synthesis of robust, constitutive, inducible, and bidirectional synthetic promoters. These promoters will enhance the transcriptional regulation of transgene expression in model plants and agricultural crops [1][2][3][4][5][6].
The Cauliflower Mosaic Virus 35S (CaMV35S) promoter has been extensively employed in plant molecular biology-based research ever since it was discovered [7][8][9][10][11][12][13][14][15][16]; still, it is regarded as a ‘dark horse’ in this field. With the gradual expansion of transgenic research, there has been an increased demand for a versatile promoter with a higher transcriptional activity. Diverse members of the family Caulimovirideae were reported to contain useful native promoters, such as the mirabilis mosaic virus (MMV) [17], figwort mosaic virus (FMV) [18], soybean chlorotic mottle soymovirus (SbCMV) [19], dahlia mosaic virus (DMV) [20], rice tungro spherical virus (RTSV) [21], tobacco vein clearing virus, (TVCV) [22], horseradish latent virus (HRLV) [23], petunia vein clearing viruses (PVCV) [24], and cassava vein mosaic virus (CVMV) [25]. Nonetheless, the available number of native promoters is insufficient to meet the increasing demand in plant biotechnology. Moreover, native promoters are neither constitutive/tissue-specific/inducible nor bidirectional in nature [26][27].
Owing to the above limitations of native promoters, researchers are engaged in designing and testing synthetic promoters capable of pursuing gene transcription in a controlled fashion, depending on the environmental stimuli. In the last two decades, redesigning promoter structure by “Cis-rearrangement” and “shuffling of sub-domains” has acquired importance in maintaining robust gene expression in plants against abiotic and biotic stresses. [10][28][29][30][31][32][33][34][35][36][37]. Such a genetically manipulated transcriptional unit is constructed by employing several state-of-the-art molecular strategies, including promoter DNA shuffling, hybridization, domain exchange, domain duplication, site-directed mutagenesis, etc. [9][10][38]. The overall purpose of these approaches is to manipulate the architecture of the Cis-element present in the promoter DNA’s backbone, in order to generate a synthetic module with altered Cis-clouding that mainly facilitates their interaction with the cognate transcription factor (TF), leading to a different module with specific functionality. Such recombinant/chimeric promoters are gaining high popularity in the plant biotechnology arena, for boosting the gene expression in plants [9].
2. Promoter Structure and Function
A promoter is the region of genomic DNA located upstream of a gene that initiates the process of transcription under specific cellular conditions. Structurally, it is modular in nature and comprises a core promoter that includes a TATA box and a CAAT box, as well as proximal and distal regions [39] (Figure 1). Functionally, promoters play a central role in regulating gene expression by their combinatorial interaction between various Cis-regulatory elements such as ‘silencers, repressors, insulators, enhancers’, and different TFs, as depicted in Figure 1 [10][40][41][42].
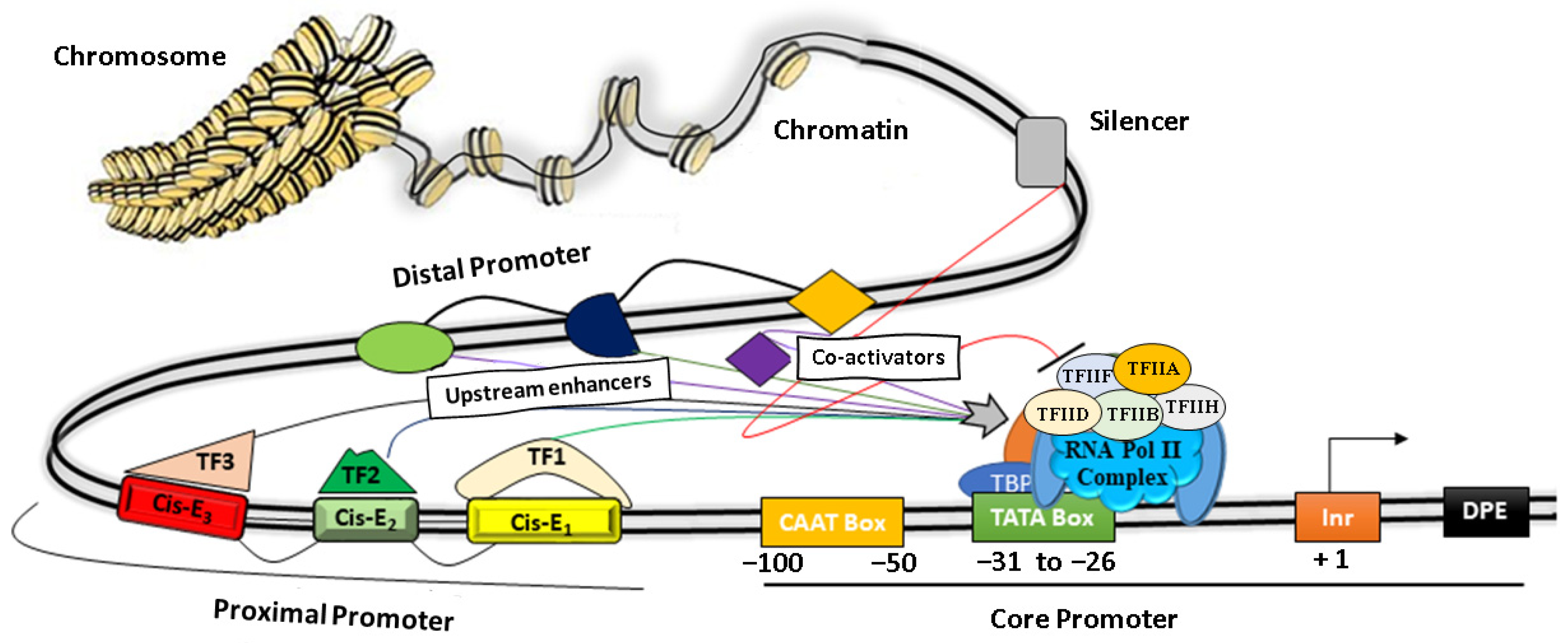
Figure 1. The basic architecture of plant promoters. The core promoter consists of a CAAT box, TATA box, Initiator element (Inr), and a downstream promoter element (DPE). Protein factors (TF IIA, IIB, IID, IIH, IIE, IIF, etc.) near the TATA box facilitate the binding of RNA Pol II to initiate transcription. The proximal regions (~1000 bp) and the distal promoter regulate the promoter activity by recruiting transcription factors (TF) on discrete Cis-elements (Cis-E).
The core promoter covers nearly 100 bp upstream of the TSS, whereas a proximal promoter extends up to a few hundred bp from the core promoter and contains several TFs binding sites [43]. The core domains consist of a conserved TATA box and motifs such as the CAAT box, GC box, Inr, and DPE. More specifically, the CAAT box recognizes and binds additional co-factors of RNA polymerase II complex such as TFIIB, TFIID, TDIIF, and TFIIE, whereas the GC box binds to various trans-activators for transcription initiations, as depicted in Figure 1. The Initiator element (Inr) is the part of the core promoter that directs the transcription initiation in typical conditions when the TATA box is dysfunctional. It has a YYANWYY consensus sequence that facilitates the binding of TF-IID and helps in strengthening the promoter [44]. Other protein factors bind to the downstream of a gene, such as Inr and regulate gene expressions [10] (Table 1, Figure 1). Several studies have shown that the proximal promoters (~1–2 kb upstream from the transcription start site) contain various important Cis-elements that strongly influence gene expression by recruiting TFs under specific cellular or environmental conditions [45][46][47]. For instance, the proximal sequence of the DREB2C promoter is sufficient for the tissue-specific spatiotemporal gene expression by heat stress in Arabidopsis [47].
Similarly, it has been shown that the simultaneous mutation at five protein binding sites (−410/−86) in the glutelin gene promoter leads to the elimination of full-length (1.8 kb) promoter activity [48]. Also, the minimal promoter proximal unit (86 to +217) of the tobacco late pollen gene g10 is sufficient for its expression in mature pollen [49]. The above studies show that the proximal regions contain crucial Cis-elements, and modulation of these elements leads to strongly altered promoter activity.
The distal promoter is the distant portion of the promoter ranging several kb upstreams of the proximal promoter and contains additional Cis-regulatory elements that have a weaker influence than the proximal promoter. Additionally, the distal regions mostly contain various Cis-elements and discrete DNA motifs (enhancers and insulators) that specifically recruit different TFs [50]. In particular, the enhancers are short DNA motifs bound with activators and they help the transcription initiation complex for enhanced gene expression. In contrast, insulators recruit repressors protein complexes to minimize or block the enhancer activity [40][51]. The downstream promoter element (DPE) recruits TF-IID at the core promoter with the help of the initiator (Inr) in the absence of the TATA box (Figure 1). Several TFs bind with Cis-motifs along with other protein factors that help the genes in enhancing/suppressing their transcriptional activity by altering the chromatin structure to facilitate/prevent the binding of the RNA polymerase complex [52][53]
Table 1. List of various Cis-motifs and trans-factors in the promoter complex.
| Promoter Cis-Motifs and Trans-Factors | Function | Reference |
|---|---|---|
| CAAT Box | Determine the efficiency of the promoter | [50] |
| GC Box | Bound by SP1 transactivator and related transcription factors | [54] |
| TATA Box | Binding site for RNA polymerase II | [50] |
| TSS/Inr | Beginning of transcription | |
| TFIIA | Promote the binding of TBP to the TATA box | |
| TFIIB | Couples to TFIID/TFIIA complex and Brings RNA polymerase II to the core promoter. | |
| TFIID | Adheres to core promoter | |
| TFIIE | Bind to the polymerase/promoter complex | |
| TFIIF | Tightly binds to the RNA Pol II | |
| TFIIH | Bind to the polymerase/promoter complex | |
| BRE | TFIIB binding sequence | |
| MTE | Motif Ten element; functions cooperatively with the Inr and is a recognition site for TFIID | [55] |
| DPE | Downstream promoter element; Recognition site for TFIID in Drosophila | [56] |
| TAFs | TBP associated factor, subunits of TFIID that assist TBP binding to DNA | [57] |
| TBP | TATA-binding protein, subunit of TFIID | [58] |
| Y Patch | Located between the TATA boxes and the TSS | [59][60] |
| CpG Island | Sp1 recognition site and simplify the regulation of gene activity by DNA-methylation | |
| I box | Light-responsive element | [61] |
| G-box | Associated with floral and root-specific expression | [60] |
| H-box | Associated with floral and root-specific expression | [60] |
| W Box | Play a role in systemic acquired resistance | [62] |
| S Box | Directs expression by fungal elicitors | [28] |
| CCAAT element | Act cooperatively with heat shock promoter elements (HSEs) | [63] |
| PB element | Potential overlap with WRKY and TGA binding | [40] |
| GCC Box | Regulation of jasmonate-responsive gene expression | [64][65] |
| HSRE | Up-regulation during the hypersensitive response | [41] |
| PC4 | Cofactor that interacts with VP16 TAD | [65] |
| SAGA | Histone acetyltransferases recruited by VP16 TAD | |
| PCAF | Play distinct roles in transactivation by MyoD | |
| UAS | Tends to contain primary regulatory elements | [66] |
3. Need of Developing Synthetic Plant Promoters
As discussed before, the major lacuna associated with the endogenous/native promoters is the weak transcriptional levels achieved within the host. Synthetic promoters offer significant ways for overcoming this obstruction and facilitating the desired gene expression, thereby allowing the efficient optimization of transgene expression at an appropriate time, place, and level [42][63]. The significant advantage of designing synthetic promoters is that it alters the promoter strength either by enhancing or reducing the promoter activity by rearranging Cis-elements via altering their copy number and spacing, which is ultimately reflected in their temporal and spatial expression pattern [27][53]. Additionally, synthetic promoters can minimize undesirable expression and remove other elements that may give rise to unwanted expression characteristics [53]. Overall, the significant role that is anticipated by synthetic promoters is to increase the target gene expression and control the unwanted and leaky expression to improve the precision and strength of the promoter [63][67]. Another objective of designing synthetic promoters is to establish sequence heterogeneity among them, which provides an added advantage for gene stacking/gene pyramiding approaches to achieve the simultaneous expression of multiple genes by avoiding promoter DNA homology-based combinations [10].
Depending upon the need of plant molecular biology, various environmental stimuli-specific synthetic promoters have been developed. [9] Researchers described the different types of synthetic plant promoters as well as the responsible host. The earlier review [9] detailed the different synthetic plant promoter types. In the present entry, the latest advancement and development of such promoters over the past decade will be discussed, along with the corresponding Cis-element and its nature; the responsible host is represented in an informative way in Table 2. Advancements made in the field of synthetic promoters are presented below.
3.1. Biotic Stress-Inducible Synthetic Promoters
The main role of a pathogen-inducible promoter is to provide resistance to a wide range of pathogens [68]. Lin et al. developed an 868-bp pathogen-inducible promoter PLsGRP1 from Lilium, which showed high activity on the pathogen and cold stress. LsGRP1 is a defense-related leaf-specific gene against a fungal pathogen causing gray mold disease. The 131-bp 3′-end region of PLsGRP1 also showed activity against various biotic, abiotic, and phytohormone exposure [69]. Four pathogen-inducible Cis-regulatory elements (PICEs) were identified in rice, namely AS-1, GCC-box, G-box, and H-box. About 53.5% of PICEs showed up- or down-regulation when exposed to pathogen attack [70].
3.2. Abiotic Stress-Inducible Synthetic Promoters
Crop productivity is majorly affected by various abiotic factors, among which high salinity and drought are the major ones. Moisture deficiency critically checks plant growth and poses constraints to crop production worldwide. This necessitates the construction of salt- and drought-inducible promoters, which can possess an optimum affinity to such stresses and can be used for imparting increased levels of tolerance to either one or both of these two abiotic stress agents. Chen et al. identified the OsHAK1 promoter sequence of 3037 nt in rice, which was found to activate under drought stress [71]. The stress-responsive gene gNAC21 was characterized from pearl millet (Pennisetum glaucum) and was reported to induce salinity tolerance in various crops [72].
3.3. Chemical-Responsive Synthetic Promoters
Certain chemicals have also been reported to regulate the transgene expression by enhanced transcription. These chemicals include various antibiotics, ethanol, herbicides, insecticides, etc. Chemically induced synthetic promoters were developed by employing probenazole, a chemical inducer which induces salicylic acid (SA) biosynthesis by inducing (SAR) systemic acquired resistance in plants [73]. A tetracycline-inducible system was developed for tobacco BY-2 suspension cells [74]. Alcohol-inducible gene expression system (AlcR-PalcA) has been successfully used in various plants such as Arabidopsis thaliana [75], Lycopersicon esculentum (tomato), and Populus sp. [76][77]. AlcR-PalcA has also been reported in microalgae Chlamydomonas reinhardtii. The alcohol-inducible AlcR-PalcA system originates from Aspergillus nidulans, a filamentous fungus [78]. A list of agrochemicals that can be used as chemical inducers is also available [79].
3.4. Development of Hormonal-Responsive Synthetic Promoters
The transcriptional regulation of different promoters is a complex process heavily influenced by several plant hormones. Wu et al. reported that the spatial pattern of abscisic acid (ABA) mediated transcriptional regulation by evaluating an ‘abscisic acid responsive element’ (ABRE) in root tissues [80]. The interaction of the synthetic promoter in association with functional genes responsible for cytosolic ABA, receptor kinase 1 (CARK1), and regulatory components of ABA receptor 11 (RCAR11) lead to drought stress tolerance [81].
3.5. Constitutive Promoters
Constitutive promoters continuously control the downstream gene expression, irrespective of space and time. To date, many promoters have been manifested with constitutive expression of genes, which have been reported to have low quality and poor yield [69]. The CaMV35SRNA promoter is the most common constitutive promoter used for various plant species, and other promoters include ubiquitin promoters [82].
3.6. Wound-Inducible Promoter
A wound-inducible promoter works exceptionally well to express the insecticidal gene in many crop plants. These promoters usually contain WRKY, W-box, and FORCA Cis-elements that recruit specific TFs for their action. Until now, several wound-inducible promoters have been isolated from different sources, namely OsDof1, Shpx6, win3.12, fib, mpiC, AoPR1, and RbPCD1 promoters [83][84][85][86][87]. Some of the above-listed promoters, such as AoPR1, mpiC1, and RbPCD1, have been used to derive CryIAc-type proteins with varying success rates [88][89]. Recently, PW220 and pRHA3B I promoters have been characterized and have been induced with wound-stress treatment in Arabidopsis [90][91]. These promoters could be used as excellent tools for developing insect-tolerant crop plants.
3.7. Bidirectional Promoters
Bidirectional promoters (BiP) are more pertinent than unidirectional promoters, as they can control the expression of two genes simultaneously, thereby saving time from expression vector construction and piling multiple genes [5]. Moreover, the limited availability of unidirectional promoters with the same expression pattern has also encouraged the application of bidirectional promoters [92]. Bidirectional promoters have been designed in many plant species such as Arabidopsis [93], rice [94], melon [95], and Capsicum annum [96], all of which have been successfully reported. In Zea mays, a novel gene stacking strategy was applied by combining a bidirectional promoter (BDP) with biCistronic approaches. This gene stacking configuration demonstrated the application of a single promoter for the coordinated expression of multiple genes in corn, a crop plant [97]. Recently, a CRISPR-Cas9 approach for genome editing in rice was investigated employing a BiP [39]. A BiP has also been used in microalga [98].
3.8. Chimeric/Hybrid Promoters
Chimeric/hybrid promoters are developed when a different functional part from one promoter is fused with the homologous/heterologous counterpart taken from another promoter. These Cis-modified promoters are developed for their utility in avoiding unpredictable and unwanted genetic recombination in plants. A new chimeric promoter was developed for controlling vascular pathogen infections by the fusion of the CaMV35S promoter and the xylogen protein 1 promoter (Px), where it serves as a vital factor in the development of the xylem in Arabidopsis [99].
3.9. Tissue-Specific Synthetic Promoters
Several tissue-specific promoters have been developed for regulating gene expression in a specific organ or tissue, such as root-specific [100], green-tissue specific [101], endosperm-specific, pollen-specific [102], etc. Green tissue-specific promoters were developed in switchgrass (Panicum virgatum L.) to enhance the biofuel production and to reduce cell wall resistance without altering the root phenotypes [103]. Recently, bidirectional green tissue-specific promoters (BiGSSP2, BiGSSP3, BiGSSP6, BiGSSP7) have been constructed [104].
Table 2. List of synthetic promoters.
| Synthetic Promoter |
Cis-Element Involved | Nature of the Promoter | Host Organism Tested |
Reference |
|---|---|---|---|---|
| SynP16 | Multiple combinations of soybean ABF, ABRE, ABRE-Like, CBF, E2F-VARIANT, G-box, GCC-Box, MYB1, MYB4, RAV1-A, and RAV1-B. Minimum 35S core promoter | Multiple expression | Glycine max | [105] |
| 12–10, 12–48, 12–79 |
Skn-1 motif, HD Zip/WUN, ABRE/boxii/Ace, CGTCA/TGACG-motif, O2 site, caat box, G box, AAGAA-motif, TATA box | Constitutive expression | Physcomitrella patens | [106] |
| AZprom (1–21) | Camv35s and Ribulose-1,5-bisphosphate carboxylase small subunit promoter | Constitutive expression | Nicotiana tabacum var Samsun | [107] |
| Saps (Sap-11) | GC content, AT and TC rich motifs, POWRS motifs | Constitutive expression | Chlamydomonasreinhardtii | [108] |
| TGA1 (CmYLCV) |
Cis-elements of CmYLCV (cestrum yellow leaf curl virus) at distal promoter region. | Constitutive expression | N. tabacum, Arabidopsis thaliana | [109] |
| SynS1, SynS2 | Various Cis-elements in the synthetic module (SynS) | Constitutive expression | Saccharum officinarum | [110] |
| Ap, Dp, ANDp | The sequences of RD29A and RD29B promoters as promoter sequences and as Cis-elements. | Inducible expression | A. thaliana | [81] |
| P_DRE:35S | Core element of camv35s promoter and tmv omega 5′-UTR, 35S core sequence. | Inducible expression | A. thaliana | [110] |
| SINC, GmubiSINC | 5′ UTR of soybean polyubiquitin promoter, Upstream camv35s | Inducible expression | Glycine max | [111] |
| SP-DDEE | e17 elements and minimal promoter, Parsley-D | Inducible expression | Brassica napus | [112] |
| GWH | JERE, SARE, GCC, 2x HSRE and 6x W-box | Inducible expression | A. thaliana | [73] |
| MAMP | Minimal promoter, uidA reporter | Microbial pathogen attack | Petroselinum crispum | [113] |
| Pmec, Pcec, Pdec and Ptec | TAM(transcription activation module), Pmec (minimum cassette expression) | Enhanced expression | Solanum lycopersicum | [16] |
| 4 x CCTC | Pi transporter 3 (StPT3) | Inducible expression under low Pi condition | Rizhophagus irregularis | [114] |
| 4 x GCC-box motifs | AtPDF1.2 promoter | Inducible expression Jasmonic acid (JA) | A. thaliana | [115] |
| ROSE 1–7 | ROSE7/GCC-box, MPK6, ERF6 | ROS response during oxidative stress | A. thaliana | [116] |
| 4 x RSRE | RSRE:;LUC reporter, GSR-motif | Rapid stress response | A. thaliana | [117] |
| PRSGA, P2RSGA, P2RSPA, PRSGPA, P2RSGPA, PR5SGPA, P2R5SGPA | ACGT and AACA motifs, Skn-1 (S), Prolam in box (P), GCN4 (G), RY repeats (R), Pzmbd1 | Seed-specific bidirectional Promoters | Zea mays | [118] |
| BiGSSP2, BiGSSP3, BiGSSP6, and BiGSSP7 | Osactin (1), Ostubulin (6, 6i) previously reported sequences, Posrbcs-550, Posrbcs-62. | Bidirectional expression in green tissues specifically. | Oryza sativa | [104] |
| SynR2 SynR1 | SynR1—Synthetic module at 5’ end SynR2—Synthetic module at 5’ and 3’end |
Root specific | N. tabacum | [119] |
| GSSP1, GSSP3, GSSP5, GSSP6, GSSP7 | First intron in rice (Act1), GT and G box. Regulatory sequences of rice, tobacco and Arabidopsis (PD 500–540, Posrbcs-550, 62 and enp3-110) |
Green tissue-specific | Oryza sativa | [120] |
| MUASMSCP | Mmvflt and mmvsgt | Constitutive expression | N. tabacum, entire plant of Petunia hybrida and protoplasts of A. thaliana | [121] |
| FSgt-PFlt, MSgt-PFlt, PFlt-UAS-2X | UAS of sub-genomic promoter transcript of MMV and FMV coupling with peanut chlorotic streak virus | Constitutive expression | N. tabacum, Amaranthus, A. thaliana, Petunia hybrida and S. lycopersicum | [35] |
| FSuasFcp, FuasFScp | Cis-elements of FMV (Figwort mosaic virus)—Sub genomic and full-length transcript promoter. | Constitutive expression | N. tabacum | [122] |
| VR-ACS1 | 5’ UTR of Vigna radiata aminocyclopropane-1-carboxylate synthase | Constitutive expression | Vigna radiata | [123] |
| p4xKST82-rd29B | rd29b: Arabidopsis rehydration responsive promoter 4xkst82: Potato KST1 promoter |
Inducible expression | N. tabacum | [124] |
| GCC2X, GCC3X, W2X, S2X | Minimal promoter of camv35s at 46 regions 2–3 copies of W, GCC and S boxes in transgenic tobacco |
Inducible expression | N. tabacum | [125] |
| EKCRM, ECCRM, EKCM | Derivative of Arabidopsis rd29a, erd1, cor15a and kin1 sequences | Inducible expression | A. thaliana | [126] |
| EFCFS-HS-(1, 2, 3) | Derived from FUAS and FS3CP containing aaag Cis-motif (Dof-1) | Inducible expression | N. tabacum | [11] |
| SP’s, SP-(EE, FF, EEFF) | Upstream regions of camv35s, F and E17 | Inducible expression | B. napus | [127] |
| pporRFP | 4XRE (regulatory element) located between B (−415, −90) and A1 (−90, −46) promoter domains of camv35s | Inducible expression | N. tabacum cv. Xanthi | [128] |
| ACGT, ACGT-2, ACGT-N-(5, 10, 25) | Single/double copies of ACGT activator motif, Pmec minimal promoter |
Inducible expression | A. thaliana and N. tabacum leaves | [30] |
| p35S-PCHS-Ω, p35S-LCHS-Ω, pOS-PCHS-Ω, pOS-LCHS-Ω |
CHSA core promoter of Petunia, CHS core promoter of Lily, Ω element, camv35s/OCS enhancer region Petunia CHSA core promoter, Lily CHS core promoter, Ω element, camv35s or OCS enhancer region |
Flower specific expression | Torenia fournieri | [129] |
| FUASCsV8CP | Up-stream activation sequence of FUAS and cp Cassava Vein Mosaic Virus | Salicyclic inducible expression | N. tabacum | [37] |
| 4D, 2S2D | 4D, 2S2D, D-box (GGAACC), Box S(AGCCACC) | Elicitor-responsive | N. tabacum | [130] |
| MSgt-FSgt | UAS domain of mmvsgt and core domain of fmvsgt | Constitutive expression | N. tabacum, A. thaliana | [131] |
| FS3-UAS-3X, FS3-UAS-2X | Fmvsgt | Constitutive expression | N. tabacum cv. | [31] |
| FS-(TGACG), FS-(TGACG)2, FS-(TGACG)3 | TGACG motif of F-Sgt | Constitutive, salicyclic acid (SA) inducible | N. tabacum and A. thaliana | [31] |
| 2x::GUS, 4x::GUS, -148-3’::GUS,-137-3’::GUS | 57 bp fragment of atmyb60 promoter and camv35s promoter | Tissue-specific expression | A. thaliana | [32] |
| pCL, pLC | CRT/DRE element (CCGAC) and TSSR of Arabidopsis cor15a promoter and potato class I promoter | Tissue-specific, cold-inducible | S. tuberosum | [132] |
| mPtDr102 | Methyl jasmonate inducible synthetic promoter derived from ptdr102 and camv35s | Bidirectional, methyl jasmonate inducible | N. tabacum | [133] |
| pOCSn-OCS, pLOCSn-OCS, pΔOCS, pLOCSΔOCS | Ocs element of octopine synthase promoter | Constitutive expression | N. tabacum | [134] |
| FUAS35SCp, MUAS35Scp | Fmvflt, mmvflt and camv35s | Bidirectional, constitutive | N. tabacum | [34] |
| A27znGlb1 | 27zn and Glb1 promoter | Tissue specific expression | Zea mays | [135] |
| 2 X W2/2 X S/2 X D, 4 X W2/4 X S | W1, W2, GCC, JERE, S, Gst1, and D boxes. CaMV35S minimal promoter | Inducible expression, Pathogen attack | Parsley | [28] |
| 2xABRE, 4xABRE | Wheat Em gene promoter | Salinity/abscisic acid inducible | Rice plant (elite Indica rice variety Khitish) | [136] |
References
- Ramesh, S.V.; Kumar, R.R.; Praveen, S. Chapter 15—Plant transcriptional regulation in modulating cross-tolerance to stress. In Priming-Mediated Stress and Cross-Stress Tolerance in Crop Plants; Hossain, M.A., Liu, F., Burritt, D.J., Fujita, M., Huang, B., Eds.; Academic Press: London, United Kingdom, 2020; pp. 231–245. ISBN 978-0-12-817892-8.
- Lehretz, G.G.; Sonnewald, S.; Hornyik, C.; Corral, J.M.; Sonnewald, U. Post-transcriptional regulation of FLOWERING LOCUS T modulates heat-dependent source-sink development in potato. Curr. Biol. 2019, 29, 1614–1624.
- de Lange, O.; Klavins, E.; Nemhauser, J. Synthetic genetic circuits in crop plants. Curr. Opin. Biotechnol. 2018, 49, 16–22.
- Nayyar, S.; Sharma, B.K.; Kaur, A.; Kalia, A.; Sanghera, G.S.; Thind, K.S.; Yadav, I.S.; Sandhu, J.S. Red rot resistant transgenic sugarcane developed through expression of β-1,3-glucanase gene. PLoS ONE 2017, 12, e0179723.
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 2016, 7, 67.
- Huang, D.; Kosentka, P.Z.; Liu, W. Synthetic biology approaches in regulation of targeted gene expression. Curr. Opin. Plant Biol. 2021, 63, 102036.
- Dong, W.; Ma, H.; Chen, C.; Li, Y. Overexpression of the OvBAN gene enhances the proanthocyanidin content in transgenic alfalfa (Medicago sativa L.). Vitr. Cell. Dev. Biol. Plant 2020, 56, 548–557.
- Kamo, K.; Thilmony, R.; Bauchan, G. Transgenic Lilium longiflorum plants containing the bar-uidA gene controlled by the rice RPC1, Agrobacterium RolD, Mas2, and CaMV 35S Promoters. Plant Cell Tissue Organ Cult. (PCTOC) 2019, 136, 303–312.
- Shrestha, A.; Khan, A.; Dey, N. Cis–Trans engineering: Advances and perspectives on customized transcriptional regulation in plants. Mol. Plant 2018, 11, 886–898.
- Dey, N.; Sarkar, S.; Acharya, S.; Maiti, I.B. Synthetic promoters in planta. Planta 2015, 242, 1077–1094.
- Ranjan, R.; Dey, N. Development of vascular tissue and stress inducible hybrid–synthetic promoters through Dof-1 Motifs rearrangement. Cell Biochem. Biophys. 2012, 63, 235–245.
- Zheng, X.; Deng, W.; Luo, K.; Duan, H.; Chen, Y.; McAvoy, R.; Song, S.; Pei, Y.; Li, Y. The cauliflower mosaic virus (CaMV) 35S promoter sequence alters the level and patterns of activity of adjacent tissue- and organ-specific gene promoters. Plant Cell Rep. 2007, 26, 1195–1203.
- Sunilkumar, G.; Mohr, L.; Lopata-Finch, E.; Emani, C.; Rathore, K.S. Developmental and tissue-specific expression of CaMV 35S promoter in cotton as revealed by GFP. Plant Mol. Biol. 2002, 50, 463–479.
- Holtorf, S.; Apel, K.; Bohlmann, H. Comparison of different constitutive and inducible promoters for the overexpression of transgenes in Arabidopsis thaliana. Plant Mol. Biol. 1995, 29, 637–646.
- Terada, R.; Shimamoto, K. Expression of CaMV35S-GUS gene in transgenic rice plants. Mol Gen. Genet. 1990, 220, 389–392.
- Koul, B.; Yadav, R.; Sanyal, I.; Sawant, S.; Sharma, V.; Amla, D.V. Cis-acting motifs in artificially synthesized expression cassette leads to enhanced transgene expression in tomato (Solanum lycopersicum L.). Plant Physiol. Biochem. 2012, 61, 131–141.
- Dey, N.; Maiti, I.B. Structure and promoter/leader deletion analysis of mirabilis mosaic virus (MMV) full-length transcript promoter in transgenic plants. Plant Mol. Biol. 1999, 40, 771–782.
- Sanger, M.; Daubert, S.; Goodman, R.M. Characteristics of a strong promoter from figwort mosaic virus: Comparison with the analogous 35s promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol. Biol. 1990, 14, 433–443.
- HU, Q.; LIU, X.; HAN, X.; LIU, Y.; JIANG, J.; XIE, Y. First detection and complete genome of soybean chlorotic mottle virus naturally infecting soybean in China by deep sequencing. J. Integr. Agric. 2019, 18, 2664–2667.
- Kuluev, B.R.; Knyazev, A.V.; Iljassowa, A.A.; Chemeris, A.V. Constitutive expression of the ARGOS gene driven by dahlia mosaic virus promoter in tobacco plants. Russ. J. Plant Physiol. 2011, 58, 507–515.
- Le, D.T.; Chu, H.D.; Sasaya, T. Creation of transgenic rice plants producing small interfering RNA of rice tungro spherical virus. GM Crops Food 2015, 6, 47–53.
- Gao, R.; Tian, Y.-P.; Wang, J.; Yin, X.; Li, X.-D.; Valkonen, J.P.T. Construction of an infectious CDNA clone and gene expression vector of tobacco vein banding mosaic virus (Genus Potyvirus). Virus Res. 2012, 169, 276–281.
- Khan, A.; Shrestha, A.; Bhuyan, K.; Maiti, I.B.; Dey, N. Structural characterization of a novel full-length transcript promoter from horseradish latent virus (HRLV) and Its transcriptional regulation by multiple stress responsive transcription factors. Plant Mol. Biol. 2018, 96, 179–196.
- Harper, G.; Richert-Pöggeler, K.R.; Hohn, T.; Hull, R. Detection of petunia vein-clearing virus: Model for the detection of DNA viruses in plants with homologous endogenous pararetrovirus sequences. J. Virol. Methods 2003, 107, 177–184.
- Verdaguer, B.; de Kochko, A.; Beachy, R.N.; Fauquet, C. Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol. Biol. 1996, 31, 1129–1139.
- Ali, S.; Kim, W.-C. A fruitful decade using synthetic promoters in the improvement of transgenic plants. Front. Plant Sci. 2019, 10, 1433.
- Liu, W.; Yuan, J.S.; Stewart, C.N., Jr. Advanced genetic tools for plant biotechnology. Nat. Rev. Genet. 2013, 14, 781–793.
- Rushton, P.J.; Reinstädler, A.; Lipka, V.; Lippok, B.; Somssich, I.E. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 2002, 14, 749–762.
- Venter, M. Synthetic promoters: Genetic control through Cis engineering. Trends Plant Sci. 2007, 12, 118–124.
- Mehrotra, R.; Mehrotra, S. Promoter activation by ACGT in response to salicylic and abscisic acids is differentially regulated by the spacing between two copies of the motif. J. Plant Physiol. 2010, 167, 1214–1218.
- Ranjan, R.; Patro, S.; Kumari, S.; Kumar, D.; Dey, N.; Maiti, I.B. Efficient chimeric promoters derived from full-length and sub-genomic transcript promoters of figwort mosaic virus (FMV). J. Biotechnol. 2011, 152, 58–62.
- Kumar, D.; Patro, S.; Ghosh, J.; Das, A.; Maiti, I.B.; Dey, N. Development of a salicylic acid inducible minimal sub-genomic transcript promoter from figwort mosaic virus with enhanced root- and leaf-activity using TGACG Motif rearrangement. Gene 2012, 503, 36–47.
- Patro, S.; Kumar, D.; Ranjan, R.; Maiti, I.B.; Dey, N. The Development of Efficient Plant Promoters for Transgene Expression Employing Plant Virus Promoters. Mol. Plant 2012, 5, 941–944.
- Patro, S.; Maiti, I.B.; Dey, N. Development of an efficient bi-directional promoter with tripartite enhancer employing three viral promoters. J. Biotechnol. 2013, 163, 311–317.
- Acharya, S.; Ranjan, R.; Pattanaik, S.; Maiti, I.B.; Dey, N. Efficient chimeric plant promoters derived from plant infecting viral promoter sequences. Planta 2014, 239, 381–396.
- Sarkar, S.; Das, A.; Khandagale, P.; Maiti, I.B.; Chattopadhyay, S.; Dey, N. Interaction of Arabidopsis TGA3 and WRKY53 transcription factors on cestrum yellow leaf curling virus (CmYLCV) promoter mediates salicylic acid-dependent gene expression in planta. Planta 2018, 247, 181–199.
- Deb, D.; Dey, N. Synthetic salicylic acid inducible recombinant promoter for translational research. J. Biotechnol. 2019, 297, 9–18.
- Nuccio, M.L.; Paul, M.; Bate, N.J.; Cohn, J.; Cutler, S.R. Where are the drought tolerant crops? An assessment of more than two decades of plant biotechnology effort in crop improvement. Plant Sci. 2018, 273, 110–119.
- Ren, Q.; Zhong, Z.; Wang, Y.; You, Q.; Li, Q.; Yuan, M.; He, Y.; Qi, C.; Tang, X.; Zheng, X.; et al. Bidirectional promoter-based CRISPR-Cas9 Systems for plant genome editing. Front. Plant Sci. 2019, 10, 1173.
- Gurr, S.J.; Rushton, P.J. Engineering plants with increased disease resistance: What are we going to express? Trends Biotechnol. 2005, 23, 275–282.
- Spitz, F.; Furlong, E.E.M. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626.
- Blazeck, J.; Alper, H.S. Promoter engineering: Recent advances in controlling transcription at the most fundamental level. Biotechnol. J. 2013, 8, 46–58.
- Smale, S.T.; Kadonaga, J.T. The RNA polymerase ii core promoter. Annu. Rev. Biochem. 2003, 72, 449–479.
- Xi, H.; Yu, Y.; Fu, Y.; Foley, J.; Halees, A.; Weng, Z. Analysis of overrepresented motifs in human core promoters reveals dual regulatory roles of YY1. Genome Res. 2007, 17, 798–806.
- Tonnessen, B.W.; Bossa-Castro, A.M.; Martin, F.; Leach, J.E. Intergenic spaces: A new frontier to improving plant health. New Phytol. 2021, 232, 1540–1548.
- Hiratsuka, T.; Makita, Y.; Yamamoto, Y.Y. Sequence-based evaluation of promoter context for prediction of transcription start sites in Arabidopsis and Rice. Sci. Rep. 2022, 12, 6976.
- Zhu, J.; Liu, M.; Liu, X.; Dong, Z. RNA polymerase II activity revealed by GRO-Seq and PNET-Seq in Arabidopsis. Nat. Plants 2018, 4, 1112–1123.
- Zheng, Z.; Kawagoe, Y.; Xiao, S.; Li, Z.; Okita, T.; Hau, T.L.; Lin, A.; Murai, N. 5′ distal and proximal cis-acting regulator elements are required for developmental control of a rice seed storage protein glutelin gene. Plant J. 1993, 4, 357–366.
- Rogers, H.J.; Bate, N.; Combe, J.; Sullivan, J.; Sweetman, J.; Swan, C.; Lonsdale, D.M.; Twell, D. Functional analysis of cis-regulatory elements within the promoter of the tobacco late pollen gene G10. Plant Mol. Biol. 2001, 45, 577–585.
- Porto, M.S.; Pinheiro, M.P.N.; Batista, V.G.L.; dos Santos, R.C.; de Albuquerque Melo Filho, P.; de Lima, L.M. Plant promoters: An approach of structure and function. Mol. Biotechnol. 2014, 56, 38–49.
- Juven-Gershon, T.; Kadonaga, J.T. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010, 339, 225–229.
- Kim, T.-K.; Shiekhattar, R. Architectural and functional commonalities between enhancers and promoters. Cell 2015, 162, 948–959.
- Rushton, P.J. What have we learned about synthetic promoter construction. In Plant Synthetic Promoters: Methods and Protocols; Hehl, R., Ed.; Springer: New York, NY, USA, 2016; pp. 1–13. ISBN 978-1-4939-6396-6.
- Bucher, P. Weight matrix descriptions of four eukaryotic RNA polymerase ii promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990, 212, 563–578.
- Deng, W.; Roberts, S.G.E. A core promoter element downstream of the TATA Box that Is recognized by TFIIB. Genes Dev. 2005, 19, 2418–2423.
- Burke, T.W.; Kadonaga, J.T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-Box-deficient promoters. Genes Dev. 1996, 10, 711–724.
- Waterworth, W.M.; Drury, G.E.; Blundell-Hunter, G.; West, C.E. Arabidopsis TAF1 Is an MRE11-interacting protein required for resistance to genotoxic stress and viability of the male gametophyte. Plant J. 2015, 84, 545–557.
- Butler, J.E.F.; Kadonaga, J.T. The RNA polymerase ii core promoter: A key component in the regulation of gene expression. Genes Dev. 2002, 16, 2583–2592.
- Yamamoto, Y.Y.; Ichida, H.; Matsui, M.; Obokata, J.; Sakurai, T.; Satou, M.; Seki, M.; Shinozaki, K.; Abe, T. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genom. 2007, 8, 67.
- Faktor, O.; Loake, G.; Dixon, R.A.; Lamb, C.J. The G-Box and H-Box in a 39 bp region of a french bean chalcone synthase promoter constitute a tissue-specific regulatory element. Plant J. 1997, 11, 1105–1113.
- Borello, U.; Ceccarelli, E.; Giuliano, G. Constitutive, Light-Responsive and Circadian Clock-Responsive Factors Compete for the Different I Box Elements in Plant Light-Regulated Promoters. Plant J. 1993, 4, 611–619.
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209.
- Liu, W.; Stewart, C.N., Jr. Plant synthetic promoters and transcription factors. Curr. Opin. Biotechnol. 2016, 37, 36–44.
- Brown, R.L.; Kazan, K.; McGrath, K.C.; Maclean, D.J.; Manners, J.M. A Role for the GCC-Box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 2003, 132, 1020–1032.
- Hirai, H.; Tani, T.; Kikyo, N. Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int. J. Dev. Biol. 2011, 54, 1589–1596.
- Fenoll, C.; Black, D.M.; Howell, S.H. The intergenic region of maize streak virus contains promoter elements involved in rightward transcription of the viral genome. EMBO J. 1988, 7, 1589–1596.
- Pandey, B.; Prakash, P.; Chandra Verma, P.; Srivastava, R. Chapter 10—Regulated gene expression by synthetic modulation of the promoter architecture in plants. In Current Developments in Biotechnology and Bioengineering; Singh, S.P., Pandey, A., Du, G., Kumar, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 235–255. ISBN 978-0-444-64085-7.
- Vijayan, J.; Devanna, B.N.; Singh, N.K.; Sharma, T.R. Cloning and functional validation of early inducible magnaporthe oryzae responsive CYP76M7 promoter from rice. Front. Plant Sci. 2015, 371.
- Lin, C.-H.; Chen, C.-Y. The pathogen-inducible promoter of defense-related LsGRP1 gene from lilium functioning in phylogenetically distinct species of plants. Plant Sci. 2017, 254, 22–31.
- Kong, W.; Ding, L.; Cheng, J.; Wang, B. Identification and expression analysis of genes with pathogen-inducible cis-regulatory elements in the promoter regions in Oryza sativa. Rice 2018, 11, 52.
- Chen, G.; Hu, J.; Lian, J.; Zhang, Y.; Zhu, L.; Zeng, D.; Guo, L.; Yu, L.; Xu, G.; Qian, Q. Functional characterization of OsHAK1 promoter in response to osmotic/drought stress by deletion analysis in transgenic rice. Plant Growth Regul. 2019, 88, 241–251.
- Shinde, H.; Dudhate, A.; Tsugama, D.; Gupta, S.K.; Liu, S.; Takano, T. Pearl millet stress-responsive NAC transcription factor PgNAC21 enhances salinity stress tolerance in Arabidopsis. Plant Physiol. Biochem. 2019, 135, 546–553.
- Zhu, Z.; Gao, J.; Yang, J.; Wang, X.; Ren, G.; Ding, Y.; Kuai, B. Synthetic promoters consisting of defined Cis-acting elements link multiple signaling pathways to probenazole-inducible system. J. Zhejiang Univ. Sci. 2015, 16, 253–263.
- Bortesi, L.; Rademacher, T.; Schiermeyer, A.; Schuster, F.; Pezzotti, M.; Schillberg, S. Development of an optimized tetracycline-inducible expression system to increase the accumulation of interleukin-10 in tobacco BY-2 suspension cells. BMC Biotechnol. 2012, 12, 40.
- Roslan, H.A.; Salter, M.G.; Wood, C.D.; White, M.R.H.; Croft, K.P.; Robson, F.; Coupland, G.; Doonan, J.; Laufs, P.; Tomsett, A.B.; et al. Characterization of the ethanol-inducible Alc gene-expression system in Arabidopsis thaliana. Plant J. 2001, 28, 225–235.
- Garoosi, G.A.; Salter, M.G.; Caddick, M.X.; Tomsett, A.B. Characterization of the ethanol-inducible Alc gene expression system in tomato. J. Exp. Bot. 2005, 56, 1635–1642.
- Filichkin, S.A.; Meilan, R.; Busov, V.B.; Ma, C.; Brunner, A.M.; Strauss, S.H. Alcohol-inducible gene expression in transgenic Populus. Plant Cell Rep. 2006, 25, 660–667.
- Lee, S.; Lee, Y.J.; Choi, S.; Park, S.-B.; Tran, Q.-G.; Heo, J.; Kim, H.-S. Development of an alcohol-inducible gene expression system for recombinant protein expression in Chlamydomonas Reinhardtii. J. Appl. Phycol. 2018, 30, 2297–2304.
- Omelina, E.S.; Yushkova, A.A.; Motorina, D.M.; Volegov, G.A.; Kozhevnikova, E.N.; Pindyurin, A.V. Optogenetic and chemical induction systems for regulation of transgene expression in plants: Use in basic and applied research. Int. J. Mol. Sci. 2022, 23, 1737.
- Wu, R.; Duan, L.; Pruneda-Paz, J.L.; Oh, D.; Pound, M.; Kay, S.; Dinneny, J.R. The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional regulation. Plant Physiol. 2018, 177, 1650–1665.
- Ge, H.; Li, X.; Chen, S.; Zhang, M.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. The expression of CARK1 or RCAR11 driven by synthetic promoters increases drought tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2018, 19, 1945.
- Honma, Y.; Yamakawa, T. High expression of GUS activities in sweet potato storage roots by sucrose-inducible minimal promoter. Plant Cell Rep. 2019, 38, 1417–1426.
- Park, S.H.; Lim, H.; Hyun, S.J.; Yun, D.W.; Yoon, U.H.; Ji, H.; Kim, T.H.; Eun, M.Y.; Kim, Y.H.; Lee, G.S. Wound-inducible expression of the OsDof1 gene promoter in a Ds insertion mutant and transgenic plants. Plant Biotechnol. Rep. 2014, 8, 305–313.
- Hollick, J.B.; Gordon, M.P. Transgenic analysis of a hybrid poplar wound-inducible promoter reveals developmental patterns of expression similar to that of storage protein genes. Plant Physiol. 1995, 109, 73–85.
- Cordero, M.J.; Raventós, D.; San Segundo, B. Expression of a maize proteinase inhibitor gene is induced in response to wounding and fungal infection: Systemic wound-response of a monocot gene. Plant J. 1994, 6, 141–150.
- Yevtushenko, D.P.; Sidorov, V.A.; Romero, R.; Kay, W.W.; Misra, S. Wound-inducible promoter from poplar is responsive to fungal infection in transgenic potato. Plant Sci. 2004, 167, 715–724.
- Perera, M.R.; Jones, M.G.K. Expression of the peroxidase gene promoter (Shpx6b) from Stylosanthes humilis in transgenic plants during insect attack. Entomol. Exp. Appl. 2004, 111, 165–171.
- Gulbitti-Onarici, S.; Zaidi, M.A.; Taga, I.; Ozcan, S.; Altosaar, I. Expression of cry1Ac in Transgenic tobacco plants under the control of a wound-inducible promoter (AoPR1) isolated from Asparagus officinalis to control Heliothis virescens and Manduca sexta. Mol. Biotechnol. 2009, 42, 341–349.
- Girijashankar, V.; Sharma, H.C.; Sharma, K.K.; Swathisree, V.; Prasad, L.S.; Bhat, B.V.; Royer, M.; Secundo, B.S.; Narasu, M.L.; Altosaar, I.; et al. Development of transgenic sorghum for insect resistance against the spotted stem borer (Chilo partellus). Plant Cell Rep. 2005, 24, 513–522.
- Pandey, S.P.; Singh, A.P.; Srivastava, S.; Chandrashekar, K.; Sane, A.P. A strong early acting wound-inducible promoter, RbPCD1pro, activates cryIAc expression within minutes of wounding to impart efficient protection against insects. Plant Biotechnol. J. 2019, 17, 1458–1470.
- Mishra, D.K.; Srivastava, R.; Pandey, B.K.; Verma, P.C.; Sawant, S.V. Identification and validation of the wound and insect bite early inducible promoter from Arabidopsis thaliana. 3 Biotech 2022, 12, 74.
- Wang, R.; Yan, Y.; Zhu, M.; Yang, M.; Zhou, F.; Chen, H.; Lin, Y. Isolation and functional characterization of bidirectional promoters in rice. Front. Plant Sci. 2016, 766.
- Banerjee, J.; Sahoo, D.K.; Dey, N.; Houtz, R.L.; Maiti, I.B. An intergenic region shared by At4g35985 and At4g35987 in Arabidopsis thaliana is a tissue specific and stress inducible bidirectional promoter analyzed in transgenic Arabidopsis and Tobacco Plants. PLoS ONE 2013, 8, e79622.
- Dhadi, S.R.; Deshpande, A.; Driscoll, K.; Ramakrishna, W. Major Cis-regulatory elements for rice bidirectional promoter activity reside in the 5′-untranslated regions. Gene 2013, 526, 400–410.
- Wang, C.; Ding, D.; Yan, R.; Yu, X.; Li, W.; Li, M. A novel bi-directional promoter cloned from melon and its activity in cucumber and tobacco. J. Plant Biol. 2008, 51, 108–115.
- Shin, R.; Kim, M.J.; Paek, K.-H. The CaTin1 (Capsicum annuum TMV-Induced Clone 1) and CaTin1-2 genes are linked head-to-head and share a bidirectional promoter. Plant Cell Physiol. 2003, 44, 549–554.
- Kumar, S.; AlAbed, D.; Whitteck, J.T.; Chen, W.; Bennett, S.; Asberry, A.; Wang, X.; DeSloover, D.; Rangasamy, M.; Wright, T.R.; et al. A combinatorial bidirectional and bicistronic approach for coordinated multi-gene expression in corn. Plant Mol. Biol. 2015, 87, 341–353.
- Poliner, E.; Takeuchi, T.; Du, Z.-Y.; Benning, C.; Farré, E.M. Nontransgenic marker-free gene disruption by an episomal CRISPR system in the Oleaginous microalga, Nannochloropsis oceanica CCMP1779. ACS Synth. Biol. 2018, 7, 962–968.
- Chen, Y.; Dong, Y.; Liang, Z.; Zhang, L.; Deng, Y. Enhanced vascular activity of a new chimeric promoter containing the full CaMV 35S promoter and the plant XYLOGEN PROTEIN 1 Promoter. 3 Biotech 2018, 8, 380.
- Li, Y.; Li, C.; Cheng, L.; Yu, S.; Shen, C.; Pan, Y. Over-expression of OsPT2 under a rice root specific promoter Os03g01700. Plant Physiol. Biochem. 2019, 136, 52–57.
- Karmakar, S.; Molla, K.A.; Chanda, P.K.; Sarkar, S.N.; Datta, S.K.; Datta, K. Green Tissue-specific co-expression of chitinase and oxalate oxidase 4 genes in rice for enhanced resistance against sheath blight. Planta 2016, 243, 115–130.
- Wang, H.; Fan, M.; Wang, G.; Zhang, C.; Shi, L.; Wei, Z.; Ma, W.; Chang, J.; Huang, S.; Lin, F. Isolation and characterization of a novel pollen-specific promoter in maize (Zea mays L.). Genome 2017, 60, 485–495.
- Liu, W.; Mazarei, M.; Ye, R.; Peng, Y.; Shao, Y.; Baxter, H.L.; Sykes, R.W.; Turner, G.B.; Davis, M.F.; Wang, Z.-Y.; et al. switchgrass (Panicum virgatum L.) Promoters for green tissue-specific expression of the MYB4 transcription factor for reduced-recalcitrance transgenic switchgrass. Biotechnol. Biofuels 2018, 11, 122.
- Bai, J.; Wang, X.; Wu, H.; Ling, F.; Zhao, Y.; Lin, Y.; Wang, R. Comprehensive construction strategy of bidirectional green tissue-specific synthetic promoters. Plant Biotechnol. J. 2020, 18, 668–678.
- Kummari, D.; Palakolanu, S.R.; Kishor, P.B.K.; Bhatnagar-Mathur, P.; Singam, P.; Vadez, V.; Sharma, K.K. An update and perspectives on the use of promoters in plant genetic engineering. J. Biosci. 2020, 45, 119.
- Peramuna, A.; Bae, H.; Rasmussen, E.K.; Dueholm, B.; Waibel, T.; Critchley, J.H.; Brzezek, K.; Roberts, M.; Simonsen, H.T. Evaluation of synthetic promoters in Physcomitrella patens. Biochem. Biophys. Res. Commun. 2018, 500, 418–422.
- Mauritz, V.; Zwiegelaar, J.P. Synthetic Promoter Construct for Transgene Expression. U.S. Patent 9,670,497, 6 June 2017.
- Scranton, M.A.; Ostrand, J.T.; Georgianna, D.R.; Lofgren, S.M.; Li, D.; Ellis, R.C.; Carruthers, D.N.; Dräger, A.; Masica, D.L.; Mayfield, S.P. Synthetic promoters capable of driving robust nuclear gene expression in the green alga Chlamydomonas reinhardtii. Algal Res. 2016, 15, 135–142.
- Sahoo, D.K.; Sarkar, S.; Maiti, I.B.; Dey, N. Novel synthetic promoters from the cestrum yellow leaf curling virus. In Plant Synthetic Promoters: Methods and Protocols; Hehl, R., Ed.; Springer: New York, NY, USA, 2016; pp. 111–138. ISBN 978-1-4939-6396-6.
- Gerasymenko, I.M.; Sheludko, Y.V. Synthetic cold-inducible promoter enhances recombinant protein accumulation during Agrobacterium-mediated transient expression in Nicotiana excelsior at chilling temperatures. Biotechnol. Lett. 2017, 39, 1059–1067.
- Grant, T.N.L.; de La Torre, C.M.; Zhang, N.; Finer, J.J. Synthetic introns help identify sequences in the 5′ UTR intron of the Glycine max polyubiquitin (Gmubi) promoter that give increased promoter activity. Planta 2017, 245, 849–860.
- Moradyar, M.; Motallebi, M.; Zamani, M.R.; Aghazadeh, R. Pathogen-induced expression of chimeric chitinase gene containing synthetic promoter confers antifungal resistance in transgenic canola. Vitr. Cell. Dev. Biol. Plant 2016, 52, 119–129.
- Kanofsky, K.; Lehmeyer, M.; Schulze, J.; Hehl, R. Analysis of microbe-associated molecular pattern-responsive synthetic promoters with the parsley protoplast system. In Plant Synthetic Promoters: Methods and Protocols; Hehl, R., Ed.; Springer: New York, NY, USA, 2016; pp. 163–174. ISBN 978-1-4939-6396-6.
- Lota, F.; Wegmüller, S.; Buer, B.; Sato, S.; Bräutigam, A.; Hanf, B.; Bucher, M. The cis-acting CTTC-P1BS module is indicative for gene function of LjVTI12, a Qb-SNARE protein gene that is required for arbuscule formation in Lotus japonicus. Plant J. 2013, 74, 280–293.
- van der Does, D.; Leon-Reyes, A.; Koornneef, A.; van Verk, M.C.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Körbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 2013, 25, 744–761.
- Wang, P.; Du, Y.; Zhao, X.; Miao, Y.; Song, C.P. The MPK6-ERF6-ROS-Responsive cis-acting element7/GCC Box complex modulates oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiol. 2013, 161, 1392–1408.
- Benn, G.; Wang, C.Q.; Hicks, D.R.; Stein, J.; Guthrie, C.; Dehesh, K. A key general stress response motif is regulated non-uniformly by CAMTA transcription factors. Plant J. 2014, 80, 82–92.
- Liu, X.; Li, S.; Yang, W.; Mu, B.; Jiao, Y.; Zhou, X.; Zhang, C.; Fan, Y.; Chen, R. Synthesis of seed-specific bidirectional promoters for metabolic engineering of anthocyanin-rich Maize. Plant Cell Physiol. 2018, 59, 1942–1955.
- Mohan, C.; Jayanarayanan, A.N.; Narayanan, S. Construction of a novel synthetic root-specific promoter and its characterization in transgenic tobacco plants. 3 Biotech 2017, 7, 234.
- Wang, R.; Zhu, M.; Ye, R.; Liu, Z.; Zhou, F.; Chen, H.; Lin, Y. Novel green tissue-specific synthetic promoters and cis-regulatory elements in rice. Sci. Rep. 2015, 5, 18256.
- Acharya, S.; Sengupta, S.; Patro, S.; Purohit, S.; Samal, S.K.; Maiti, I.B.; Dey, N. Development of an intra-molecularly shuffled efficient chimeric plant promoter from plant infecting mirabilis mosaic virus promoter sequence. J. Biotechnol. 2014, 169, 103–111.
- Ranjan, R.; Patro, S.; Pradhan, B.; Kumar, A.; Maiti, I.B.; Dey, N. Development and functional analysis of novel genetic promoters using dna shuffling, hybridization and a combination thereof. PLoS ONE 2012, 7, e31931.
- Wever, W.; McCallum, E.J.; Chakravorty, D.; Cazzonelli, C.I.; Botella, J.R. The 5′ untranslated region of the VR-ACS1 MRNA acts as a strong translational enhancer in plants. Transgenic Res. 2010, 19, 667–674.
- Na, J.-K.; Metzger, J.D. Chimeric promoter mediates guard cell-specific gene expression in tobacco under water deficit. Biotechnol. Lett. 2014, 36, 1893–1899.
- Yeri, S.B.; Bhat, R.S.; Kuruvinashetti, M.S. Functional analysis of synthetic promoters containing pathogen-responsive cis-elements. Mol. Plant Breed. 2013, 4, 270–276.
- Hou, L.; Chen, L.; Wang, J.; Xu, D.; Dai, L.; Zhang, H.; Zhao, Y. Construction of stress responsive synthetic promoters and analysis of their activity in transgenic Arabidopsis thaliana. Plant Mol. Biol. Rep. 2012, 30, 1496–1506.
- Shokouhifar, F.; Zamani, M.R.; Motallebi, M.; Mousavi, A.; Malboobi, M.A. Construction and functional analysis of pathogen-inducible synthetic promoters in Brassica napus. Biol. Plant. 2011, 55, 689.
- Liu, W.; Mazarei, M.; Rudis, M.R.; Fethe, M.H.; Stewart, C.N., Jr. Rapid in vivo analysis of synthetic promoters for plant pathogen phytosensing. BMC Biotechnol. 2011, 11, 108.
- Du, L.; Lou, Q.; Zhang, X.; Jiao, S.; Liu, Y.; Wang, Y. Construction of flower-specific chimeric promoters and analysis of their activities in transgenic torenia. Plant Mol. Biol. Rep. 2014, 32, 234–245.
- Niemeyer, J.; Ruhe, J.; Machens, F.; Stahl, D.J.; Hehl, R. Inducible expression of P50 from TMV for increased resistance to bacterial crown gall disease in tobacco. Plant Mol. Biol. 2014, 84, 111–123.
- Kumar, D.; Patro, S.; Ranjan, R.; Sahoo, D.K.; Maiti, I.B.; Dey, N. Development of useful recombinant promoter and its expression analysis in different plant cells using confocal laser scanning microscopy. PLoS ONE 2011, 6, e24627.
- Li, M.; Song, B.; Zhang, Q.; Liu, X.; Lin, Y.; Ou, Y.; Zhang, H.; Liu, J. A synthetic tuber-specific and cold-induced promoter is applicable in controlling potato cold-induced sweetening. Plant Physiol. Biochem. 2013, 67, 41–47.
- Zheng, H.; Lei, Y.; Lin, S.; Zhang, Q.; Zhang, Z. Bidirectionalization of a methyl jasmonate-inducible plant promoter. Biotechnol. Lett. 2011, 33, 387–393.
- Liu, S.; Bao, Y. Effects of copy number of an octopine synthase enhancer element and its distance from the TATA Box on heterologous expression in transgenic tobacco. Acta Physiol. Plant. 2009, 31, 705–710.
- Shepherd, C.T.; Scott, M.P. Construction and evaluation of a maize (Zea mays) chimaeric promoter with activity in kernel endosperm and embryo. Biotechnol. Appl. Biochem. 2009, 52, 233–243.
- Ganguly, M.; Roychoudhury, A.; Sarkar, S.N.; Sengupta, D.N.; Datta, S.K.; Datta, K. Inducibility of three salinity/AbsCisic acid-regulated promoters in transgenic rice with GusA reporter gene. Plant Cell Rep. 2011, 30, 1617–1625.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.0K
Revisions:
2 times
(View History)
Update Date:
28 Apr 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




