Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Guolong Zhang | -- | 1120 | 2023-04-17 15:46:35 | | | |
| 2 | Sirius Huang | Meta information modification | 1120 | 2023-04-18 04:06:00 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Tobin, I.; Zhang, G. Host Defense Peptides. Encyclopedia. Available online: https://encyclopedia.pub/entry/43123 (accessed on 07 February 2026).
Tobin I, Zhang G. Host Defense Peptides. Encyclopedia. Available at: https://encyclopedia.pub/entry/43123. Accessed February 07, 2026.
Tobin, Isabel, Guolong Zhang. "Host Defense Peptides" Encyclopedia, https://encyclopedia.pub/entry/43123 (accessed February 07, 2026).
Tobin, I., & Zhang, G. (2023, April 17). Host Defense Peptides. In Encyclopedia. https://encyclopedia.pub/entry/43123
Tobin, Isabel and Guolong Zhang. "Host Defense Peptides." Encyclopedia. Web. 17 April, 2023.
Copy Citation
Host defense peptides (HDPs), also known as antimicrobial peptides, are an important component of the innate immune system. HDPs possess both antimicrobial and immunomodulatory properties. HDPs and their derivatives are being actively explored for antimicrobial therapies. A host-directed approach to stimulate the synthesis of endogenous HDPs is also being developed to treat infections with a minimum risk for developing antimicrobial resistance.
host defense peptides
antimicrobial resistance
antimicrobial peptides
antibiotic alternatives
1. Introduction
Antimicrobial resistance has emerged as a major world health concern over past decades [1]. Excessive usage of antibiotics in human medicine and agriculture production has driven the emergence of resistant bacterial strains [1]. This issue has led to a significant increase in deaths associated with infections that were once treatable with antibiotics [1]. Development of novel antibiotics and antimicrobial approaches has become an intensive area of research [2]. Host defense peptides (HDPs), also known as antimicrobial peptides, elicit strong antimicrobial activity and regulate the immune response to protect against infections while minimizing inflammation [3][4]. As an important chemical barrier of the innate immune system, HDPs are small peptides (approximately 15–45 amino acids) found in virtually all organisms including mammals, birds, reptiles, amphibians, plants, and fungi [3][4][5].
2. Classifications of HDPs
Two major HDP families in vertebrate animals are cathelicidins and defensins [3][4]. Cathelicidins are produced abundantly in myeloid cells such as neutrophils and monocytes as well as skin keratinocytes and epithelial cells lining the respiratory, gastrointestinal, and urogenital tracts [6]. Synthesized initially as a precursor, cathelicidins are proteolytically cleaved from the cathelin pro-segment to become biologically active and released upon activation [6]. Defensins are characterized by the presence of six cysteine residues in defined positions [7]. Based on the location and spacing pattern of the cysteines, defensins are further divided into three subfamilies, namely α-, β-, and θ-defensins [7]. The α-defensins are produced abundantly by neutrophils and intestinal Paneth cells, while β-defensins are produced largely by the skin keratinocytes and epithelial cells of the gastrointestinal and urogenital tracts [7]. On the other hand, θ-defensins have a unique cyclic structure and have only been found in primates [8]. Like cathelicidins, α- and θ-defensins are translated as inactive precursors that are proteolytically cleaved at highly conserved sites to become mature active peptides to be stored in the granules of neutrophils or Paneth cells [8].
3. Mechanism of Antimicrobial Action of HDPs
HDPs are active against both Gram-positive and Gram-negative bacteria, fungi, parasites, and enveloped viruses, as well as cancer cells [3][4]. Several models, such as barrel stave, carpet, and toroidal pore, have been proposed to explain the antibacterial activity of HDPs [3][4] (Figure 1). The most accepted mechanism is based on the cationic and amphipathic natures of HDPs, which allow these peptides to interact with the outer membrane of bacteria composed of negatively charged phospholipid head groups. The cationicity of HDPs drives their attraction to the bacterial membrane causing disruption of the membrane structure leading to the leakage of cellular contents and bacterial lysis [3][4]. Additionally, other antibacterial mechanisms have been revealed with several HDPs (Figure 1). For example, human α-defensin 6 (HD6) forms a nanonet structure to entrap bacteria to prevent translocation into intestinal epithelium [9]. Proline-rich HDPs, such as indolicidin, kill bacteria by binding to intracellular targets such as DNA without membrane lysis [10]. Proline-arginine rich PR-39 directly inhibits or degrades proteins involved in DNA replication in the bacterial nucleus [11]. Furthermore, human β-defensin 3 (HBD3), has been shown to inhibit bacterial cell wall synthesis by interacting with the cell wall building block, lipid II [12].
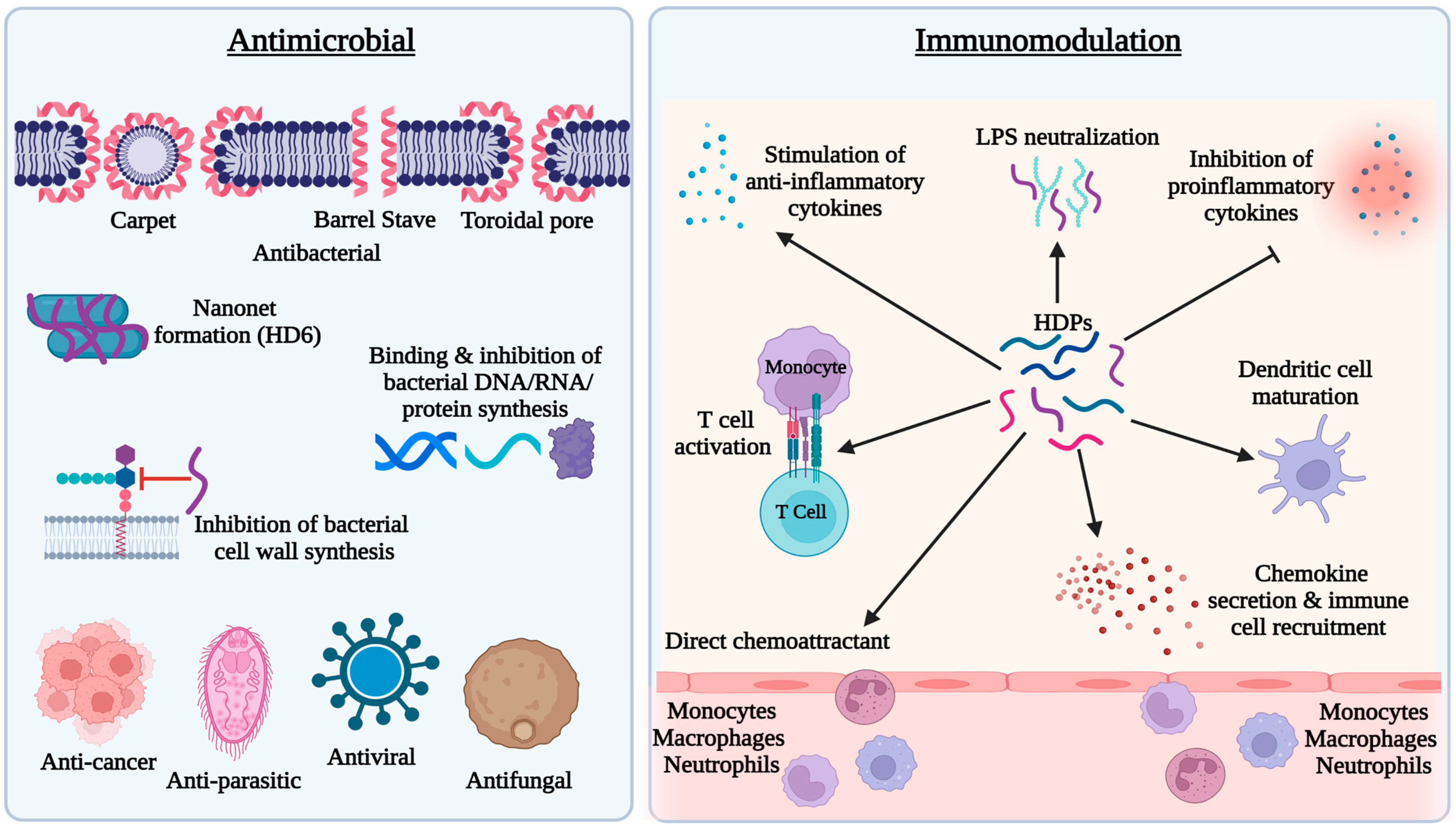
Figure 1. Antimicrobial and immunomodulatory activities of host defense peptides (HDPs). HDPs possess direct antimicrobial activity against bacteria, fungi, viruses, and parasites mainly through membrane disruption. A few HDPs also inhibit bacteria by forming nanonets or targeting bacterial DNA, RNA, proteins, or lipid II of the cell wall. Immunomodulatory effects of HDPs mainly include recruitment and activation of immune cells and regulation of inflammatory response.
4. Role of HDPs in Innate and Adaptive Immunity
In addition to direct antimicrobial activity, HDPs possess immunomodulatory activities [3][6] (Figure 1). HDPs are capable of directly recruiting different types of immune cells to the site of infection or indirectly through stimulation of chemokine production. For example, human cathelicidin LL-37 is chemotactic for neutrophils, monocytes, and T lymphocytes through engaging formyl peptide receptor 2 [13], while human β-defensins such as HBD2 and HBD3 chemoattract neutrophils and monocytes through interacting with chemokine receptor CCR2 [14] and they recruit dendritic cells and T cells through CCR6 [15]. Additionally, HDPs such as LL-37 and HBD3 can induce the expression of a variety of chemokines, such as CCL2/monocyte chemoattractant protein-1, CCL3/macrophage inflammatory protein-1α, CCL4/macrophage inflammatory protein-1β, CXCL1/Gro-α, and CCL22/macrophage-derived chemokine, which attract monocytes, macrophages, neutrophils, dendritic cells, and T cells subsequently to the sites of inflammation [16]. In addition to chemoattraction, many HDPs further induce maturation and activation of the cells involved in innate and adaptive immunity. For example, β-defensins induce the expression of co-stimulatory molecules, CD80, CD86, and CD40, on monocytes and dendritic cells in a Toll-like receptor-dependent manner [17][18].
Although they are capable of eliciting proinflammatory and immune responses to facilitate the clearance of pathogens, HDPs mediate largely anti-inflammatory responses during inflammation and infection by inhibiting the expression of proinflammatory cytokines such as interleukin (IL)-1β, IL-6, and tumor necrosis factor-α (TNF-α), while stimulating the production of anti-inflammatory cytokines such as IL-10 [19][20] (Figure 1). Furthermore, cationic HDPs also bind to anionic lipopolysaccharides (LPS) to neutralize its effect on inflammation [3][6].
5. Transcriptional Regulation of HDPs
As an important component of innate defense, certain HDPs such as human α-defensins and HBD1 are constitutively produced, while other HDPs like HBD2, HBD3, and HBD4 are induced upon infection and injury [21][22]. Pathogen-associated molecular patterns (PAMPs), such as LPS, bacterial DNA, and flagellin, can induce the expression of HDPs [21]. For example, Psuedomonas aeruginosa rhamnolipids act as a PAMP to activate transcription factors, such as nuclear factor κB (NF-κB) and activator protein-1 (AP-1), causing induction of HBD2 in keratinocytes [23][24]. Proinflammatory cytokines such as TNF-α and IL-1α are known to induce the expression of HDPs such as HBD2 [25].
Additionally, a diverse group of small-molecule compounds have been shown to induce expression of HDPs [26][27][28][29]. Enhancing HDP synthesis by short-chain fatty acids (SCFAs) such as acetate, propionate, and butyrate has been well documented [29]. Medium-chain fatty acids are also capable of regulating HDP synthesis, albeit with reduced efficacy [30][31]. Amino acids such as L-arginine, L-isoleucine, leucine, and valine can induce β-defensin synthesis in the intestinal epithelium [32]. Histone deacetylase (HDAC) inhibitors, such as entinostat, valproic acid, and trichostatin, are very effective at upregulating HDP synthesis [28]. Other compounds such as vitamin D3, zinc, β-glucan, fructan, and lactose are also capable of inducing HDP synthesis [27]. Furthermore, probiotic bacteria such as Lactobacillus strains have the ability to upregulate β-defensin synthesis as well [33]. Recent development of several high-throughput screening assays has led to identification of a number of HDP-inducing compounds and their efficacy in disease control and prevention is being characterized [34][35][36][37][38][39].
References
- Watkins, R.R.; Bonomo, R.A. Overview: The Ongoing Threat of Antimicrobial Resistance. Infect. Dis. Clin. N. Am. 2020, 34, 649–658.
- Schrader, S.M.; Vaubourgeix, J.; Nathan, C. Biology of antimicrobial resistance and approaches to combat it. Sci. Transl. Med. 2020, 12, eaaz6992.
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332.
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230.
- Thakur, A.; Sharma, A.; Alajangi, H.K.; Jaiswal, P.K.; Lim, Y.B.; Singh, G.; Barnwal, R.P. In pursuit of next-generation therapeutics: Antimicrobial peptides against superbugs, their sources, mechanism of action, nanotechnology-based delivery, and clinical applications. Int. J. Biol. Macromol. 2022, 218, 135–156.
- Alford, M.A.; Baquir, B.; Santana, F.L.; Haney, E.F.; Hancock, R.E.W. Cathelicidin Host Defense Peptides and Inflammatory Signaling: Striking a Balance. Front. Microbiol. 2020, 11, 1902.
- Xu, D.; Lu, W. Defensins: A Double-Edged Sword in Host Immunity. Front. Immunol. 2020, 11, 764.
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557.
- Chu, H.; Pazgier, M.; Jung, G.; Nuccio, S.P.; Castillo, P.A.; de Jong, M.F.; Winter, M.G.; Winter, S.E.; Wehkamp, J.; Shen, B.; et al. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 2012, 337, 477–481.
- Gennaro, R.; Zanetti, M.; Benincasa, M.; Podda, E.; Miani, M. Pro-rich antimicrobial peptides from animals: Structure, biological functions and mechanism of action. Curr. Pharm. Des. 2002, 8, 763–778.
- Cardoso, M.H.; Meneguetti, B.T.; Costa, B.O.; Buccini, D.F.; Oshiro, K.G.N.; Preza, S.L.E.; Carvalho, C.M.E.; Migliolo, L.; Franco, O.L. Non-Lytic Antibacterial Peptides That Translocate Through Bacterial Membranes to Act on Intracellular Targets. Int. J. Mol. Sci. 2019, 20, 4877.
- Sass, V.; Schneider, T.; Wilmes, M.; Körner, C.; Tossi, A.; Novikova, N.; Shamova, O.; Sahl, H.-G. Human beta-Defensin 3 Inhibits Cell Wall Biosynthesis in Staphylococci. Infect. Immun. 2010, 78, 2793–2800.
- Yang, D.; Chen, Q.; Schmidt, A.P.; Anderson, G.M.; Wang, J.M.; Wooters, J.; Oppenheim, J.J.; Chertov, O. Ll-37, the Neutrophil Granule–And Epithelial Cell–Derived Cathelicidin, Utilizes Formyl Peptide Receptor–Like 1 (Fprl1) as a Receptor to Chemoattract Human Peripheral Blood Neutrophils, Monocytes, and T Cells. J. Exp. Med. 2000, 192, 1069–1074.
- Rohrl, J.; Yang, D.; Oppenheim, J.J.; Hehlgans, T. Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J. Immunol. 2010, 184, 6688–6694.
- Yang, D.; Chertov, O.; Bykovskaia, S.N.; Chen, Q.; Buffo, M.J.; Shogan, J.; Anderson, M.; Schroder, J.M.; Wang, J.M.; Howard, O.M.; et al. Beta-defensins: Linking innate and adaptive immunity through dendritic and T cell CCR6. Science 1999, 286, 525–528.
- Petrov, V.; Funderburg, N.; Weinberg, A.; Sieg, S. Human β defensin-3 induces chemokines from monocytes and macrophages: Diminished activity in cells from HIV-infected persons. Immunology 2013, 140, 413–420.
- Biragyn, A.; Ruffini, P.A.; Leifer, C.A.; Klyushnenkova, E.; Shakhov, A.; Chertov, O.; Shirakawa, A.K.; Farber, J.M.; Segal, D.M.; Oppenheim, J.J.; et al. Toll-Like Receptor 4-Dependent Activation of Dendritic Cells by β-Defensin 2. Science 2002, 298, 1025–1029.
- Funderburg, N.; Lederman, M.M.; Feng, Z.; Drage, M.G.; Jadlowsky, J.; Harding, C.V.; Weinberg, A.; Sieg, S.F. Human -defensin-3 activates professional antigen-presenting cells via Toll-like receptors 1 and 2. Proc. Natl. Acad. Sci. USA 2007, 104, 18631–18635.
- Liu, H.; Yu, H.; Gu, Y.; Xin, A.; Zhang, Y.; Diao, H.; Lin, D. Human beta-defensin DEFB126 is capable of inhibiting LPS-mediated inflammation. Appl. Microbiol. Biotechnol. 2013, 97, 3395–3408.
- Mookherjee, N.; Brown, K.L.; Bowdish, D.M.E.; Doria, S.; Falsafi, R.; Hokamp, K.; Roche, F.M.; Mu, R.; Doho, G.H.; Pistolic, J.; et al. Modulation of the TLR-Mediated Inflammatory Response by the Endogenous Human Host Defense Peptide LL-37. J. Immunol. 2006, 176, 2455–2464.
- Contreras, G.; Shirdel, I.; Braun, M.S.; Wink, M. Defensins: Transcriptional regulation and function beyond antimicrobial activity. Dev. Comp. Immunol. 2020, 104, 103556.
- Lyu, W.; Curtis, A.R.; Sunkara, L.T.; Zhang, G. Transcriptional Regulation of Antimicrobial Host Defense Peptides. Curr. Protein Pept. Sci. 2015, 16, 672–679.
- Wehkamp, K.; Schwichtenberg, L.; Schröder, J.-M.M.; Harder, J. Pseudomonas aeruginosa- and IL-1β-Mediated Induction of Human β-Defensin-2 in Keratinocytes Is Controlled by NF-κB and AP-1. J. Investig. Dermatol. 2006, 126, 121–127.
- Gerstel, U.; Czapp, M.; Bartels, J.; Schröder, J.-M. Rhamnolipid-induced shedding of flagellin from Pseudomonas aeruginosa provokes hBD-2 and IL-8 response in human keratinocytes. Cell. Microbiol. 2009, 11, 842–853.
- O’Neil, D.A.; Porter, E.M.; Elewaut, D.; Anderson, G.M.; Eckmann, L.; Ganz, T.; Kagnoff, M.F. Expression and Regulation of the Human β-Defensins hBD-1 and hBD-2 in Intestinal Epithelium. J. Immunol. 1999, 163, 6718–6724.
- Bergman, P.; Raqib, R.; Rekha, R.S.; Agerberth, B.; Gudmundsson, G.H. Host Directed Therapy Against Infection by Boosting Innate Immunity. Front. Immunol. 2020, 11, 1209.
- Chen, J.; Zhai, Z.; Long, H.; Yang, G.; Deng, B.; Deng, J. Inducible expression of defensins and cathelicidins by nutrients and associated regulatory mechanisms. Peptides 2020, 123, 170177.
- Rodriguez-Carlos, A.; Jacobo-Delgado, Y.M.; Santos-Mena, A.O.; Rivas-Santiago, B. Modulation of cathelicidin and defensins by histone deacetylase inhibitors: A potential treatment for multi-drug resistant infectious diseases. Peptides 2021, 140, 170527.
- Robinson, K.; Ma, X.; Liu, Y.; Qiao, S.; Hou, Y.; Zhang, G. Dietary modulation of endogenous host defense peptide synthesis as an alternative approach to in-feed antibiotics. Anim. Nutr. 2018, 4, 160–169.
- Jiang, W.; Sunkara, L.T.; Zeng, X.; Deng, Z.; Myers, S.M.; Zhang, G. Differential regulation of human cathelicidin LL-37 by free fatty acids and their analogs. Peptides 2013, 50, 129–138.
- Sunkara, L.T.; Jiang, W.; Zhang, G. Modulation of antimicrobial host defense peptide gene expression by free fatty acids. PLoS ONE 2012, 7, e49558.
- Ren, M.; Zhang, S.; Liu, X.; Li, S.; Mao, X.; Zeng, X.; Qiao, S. Different Lipopolysaccharide Branched-Chain Amino Acids Modulate Porcine Intestinal Endogenous β-Defensin Expression through the Sirt1/ERK/90RSK Pathway. J. Agric. Food Chem. 2016, 64, 3371–3379.
- Schlee, M.; Harder, J.; Koten, B.; Stange, E.F.; Wehkamp, J.; Fellermann, K. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clin. Exp. Immunol. 2008, 151, 528–535.
- Lyu, W.; Deng, Z.; Zhang, G. High-Throughput Screening for Epigenetic Compounds That Induce Human beta-Defensin 1 Synthesis. Antibiotics 2023, 12, 186.
- Lyu, W.; Mi, D.; Vinson, P.N.; Zhang, G. Large-scale Identification of Multiple Classes of Host Defense Peptide-Inducing Compounds for Antimicrobial Therapy. Int. J. Mol. Sci. 2022, 23, 8400.
- Deng, Z.; Lyu, W.; Zhang, G. High-Throughput Identification of Epigenetic Compounds to Enhance Chicken Host Defense Peptide Gene Expression. Antibiotics 2022, 11, 933.
- Wang, J.; Lyu, W.; Zhang, W.; Chen, Y.; Luo, F.; Wang, Y.; Ji, H.; Zhang, G. Discovery of natural products capable of inducing porcine host defense peptide gene expression using cell-based high throughput screening. J. Anim. Sci. Biotechnol. 2021, 12, 14.
- Lyu, W.; Deng, Z.; Sunkara, L.T.; Becker, S.; Robinson, K.; Matts, R.; Zhang, G. High Throughput Screening for Natural Host Defense Peptide-Inducing Compounds as Novel Alternatives to Antibiotics. Front. Cell. Infect. Microbiol. 2018, 8, 191.
- Nylen, F.; Miraglia, E.; Cederlund, A.; Ottosson, H.; Stromberg, R.; Gudmundsson, G.H.; Agerberth, B. Boosting innate immunity: Development and validation of a cell-based screening assay to identify LL-37 inducers. Innate Immun. 2014, 20, 364–376.
More
Information
Subjects:
Immunology; Infectious Diseases
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
994
Revisions:
2 times
(View History)
Update Date:
18 Apr 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




