
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Louis Broussard | -- | 2324 | 2023-04-11 18:00:50 | | | |
| 2 | Louis Broussard | + 9 word(s) | 2333 | 2023-04-11 18:07:03 | | | | |
| 3 | Lindsay Dong | -10 word(s) | 2323 | 2023-04-13 07:46:55 | | | | |
| 4 | Lindsay Dong | -4 word(s) | 2319 | 2023-04-13 07:48:08 | | |
Video Upload Options
Phloem sap transport is essential for plant nutrition and development since it mediates redistribution of nutrients, metabolites and signaling molecules. However, its biochemical composition is not so well-known because phloem sap sampling is difficult and does not always allow extensive chemical analysis. Efforts have been devoted to metabolomics analyses of phloem sap using either liquid chromatography or gas chromatography coupled with mass spectrometry. Phloem sap metabolomics is of importance to understand how metabolites can be exchanged between plant organs and how metabolite allocation may impact plant growth and development.
1. Introduction
2. Phloem Sap Metabolites
3. Metabolic Pathways Reflected by Phloem Sap Metabolome
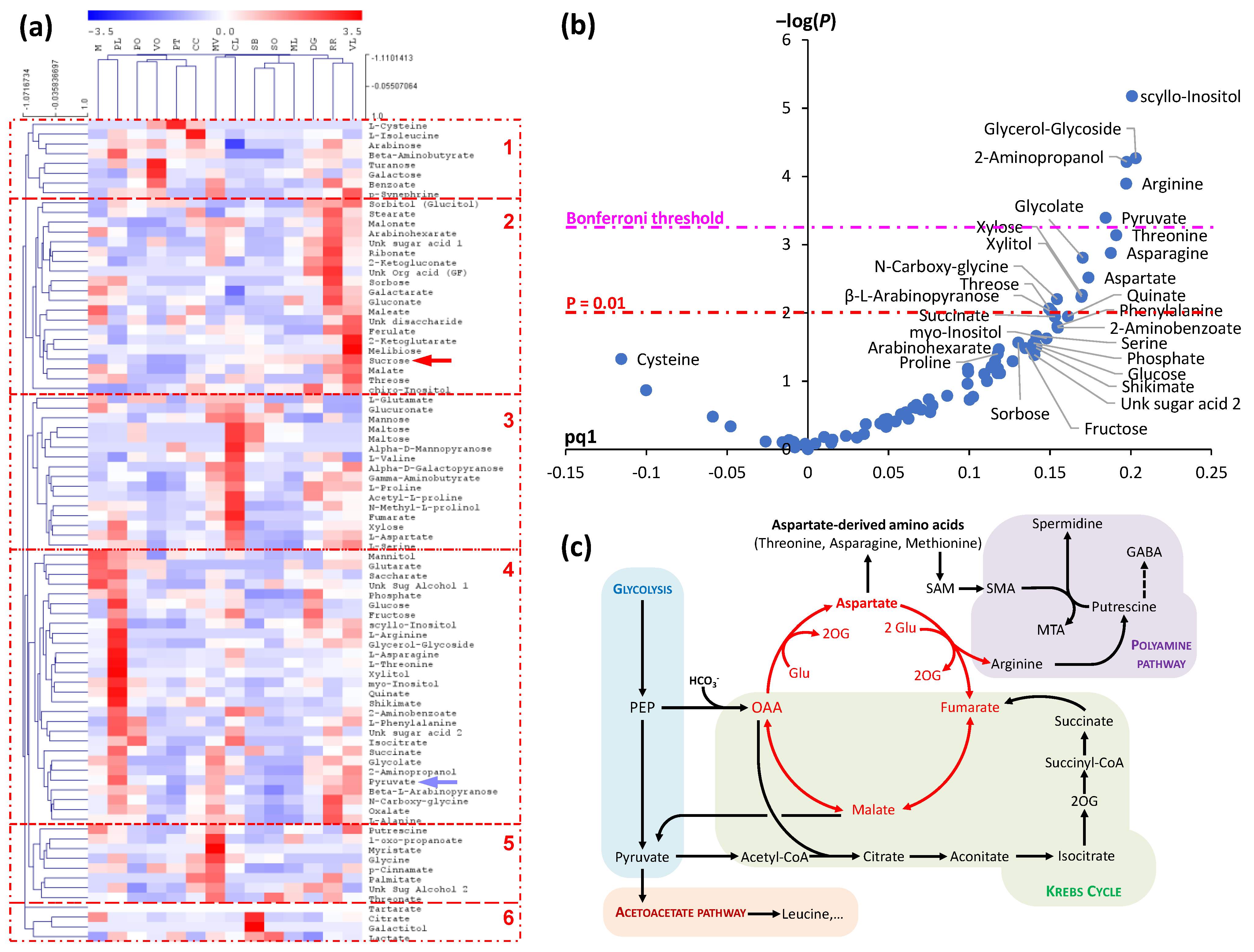
4. Phloem Sap Metabolites and Plant Resistance to Environmental Cues
Amino acid composition in phloem sap appears to respond to environmental factors such as nutrient availability. When supplied with high levels of nitrogen (N), Glu content increase in Arabidopsis phloem sap and conversely, Pro, Gln and γ-aminobutyrate (4-aminobutyrate; GABA) increase under low N availability [31]. Total amino acid concentration also changes with N supply as has been shown in canola [53]. Other conditions such as low phosphorus (P) availability and water deficit also affect amino acid composition [31][54]. Polyol content in phloem sap (such as sorbitol and erythritol) have also been found to impact drought and cold tolerance [24][28]. Recently, metabolomics analyses of cucumber plants subjected to phosphorus deficiency have shown important changes in phloem sap metabolome, not only in carbohydrates (galactitol, fructose) but also in organic acids (e.g., oxalate, citrate, fatty acids) along with nitrogenous compounds (e.g., ethanolamine, 4-aminobutyrate and pyroglutamate) [33]. Metabolomics analyses of phloem sap exudates during root waterlogging have found a change in the sugar to organic acid ratio, suggesting (i) a crucial role of the balance between loading in shoots and unloading in roots for sap composition and (ii) potentially, a negative feedback of metabolite accumulation in phloem sap onto shoot metabolism, such as sugar interconversions and respiration [55].
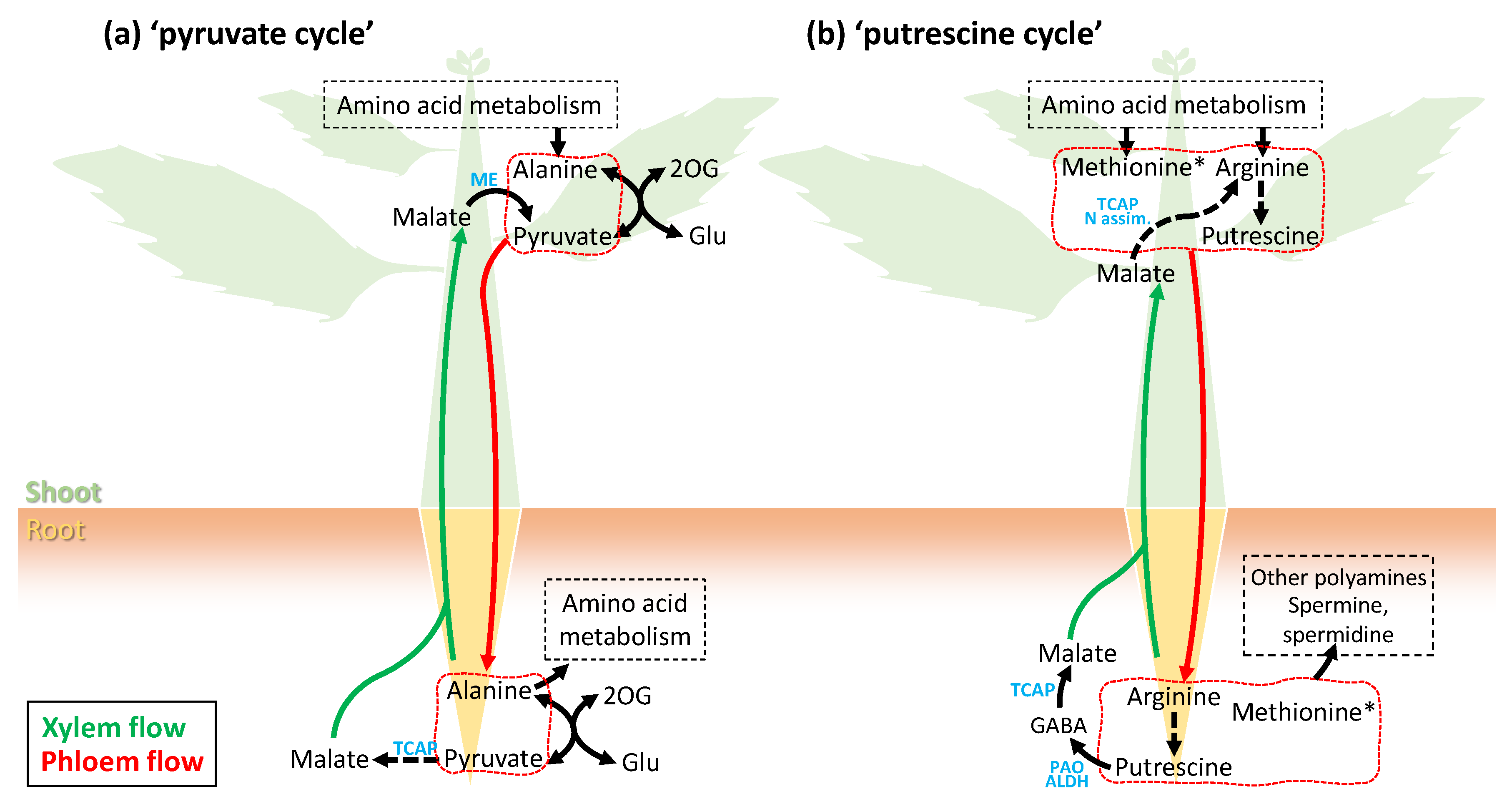
5. Phloem Sap Metabolome: Unforeseen Whole-Plant Metabolic Cycles?
References
- Dinant, S.; Lemoine, R. The phloem pathway: New issues and old debates. Comptes Rendus Biol. 2010, 333, 307–319.
- Evert, R.F. Phloem Structure and Histochemistry. Annu. Rev. Plant Physiol. 1977, 28, 199–222.
- Cronshaw, J. Phloem Structure and Function. Annu. Rev. Plant Physiol. 1981, 32, 465–484.
- Golecki, B.; Schulz, A.; Thompson, G.A. Translocation of Structural P Proteins in the Phloem. Plant Cell 1999, 11, 127–140.
- Ernst, A.M.; Jekat, S.B.; Zielonka, S.; Muller, B.; Neumann, U.; Ruping, B.; Twyman, R.M.; Krzyzanek, V.; Prufer, D.; Noll, G.A. Sieve element occlusion (SEO) genes encode structural phloem proteins involved in wound sealing of the phloem. Proc. Natl. Acad. Sci. USA 2012, 109, E1980–E1989.
- Sellier, D.; Harrington, J.J. Phloem transport in trees: A generic surface model. Ecol. Model. 2014, 290, 102–109.
- Holbrook, N.M.; Knoblauch, M. Editorial overview: Physiology and metabolism: Phloem: A supracellular highway for the transport of sugars, signals, and pathogens. Curr. Opin. Plant Biol. 2018, 43, iii–vii.
- Weiler, E.W. Determination of phytohormones in phloem exudate from tree species by radioimmunoassay. Planta 1981, 152, 168–170.
- Simpson, R.J. Nitrogen Redistribution during Grain Growth in Wheat (Triticum aestivum L.) 1: IV. Development of a Quantitative Model of the Translocation of Nitrogen to the Grain. Plant Physiol. 1983, 71, 7–14.
- Rennenberg, H. Analysis of uptake and allocation of nitrogen and sulphur compounds by trees in the field. J. Exp. Bot. 1996, 47, 1491–1498.
- Anderson, J.W. Sulphur Distribution and Redistribution in Vegetative and Generative Plants. In Sulphur in Plants; Springer: Dordrecht, The Netherlands, 2003; pp. 113–134.
- Youssefi, F.; Brown, P.; Weinbaum, S. Relationship between tree nitrogen status, xylem and phloem sap amino acid concentrations, and apparent soil nitrogen uptake by almond trees (Prunus dulcis). J. Hortic. Sci. Biotechnol. 2015, 75, 62–68.
- Pate, J.S.; Jeschke, W.D. Mineral uptake and transport in xylem and phloem of the proteaceous tree, Banksia prionotes. Plant Soil 1993, 155, 273–276.
- Lohaus, G. Review primary and secondary metabolites in phloem sap collected with aphid stylectomy. J. Plant Physiol. 2022, 271, 153645.
- Ohshima, T.; Hayashi, H.; Chino, M. Collection and Chemical Composition of Pure Phloem Sap from Zea mays L. Plant Cell Physiol. 1990, 31, 735–737.
- Lohaus, G.; Moellers, C. Phloem transport of amino acids in two Brassica napus L. genotypes and one B. carinata genotype in relation to their seed protein content. Planta 2000, 211, 833–840.
- Hayashi, H.; Chino, M. Chemical Composition of Phloem Sap from the Uppermost Internode of the Rice Plant. Plant Cell Physiol. 1990, 31, 247–251.
- Winter, H.; Lohaus, G.; Heldt, H.W. Phloem Transport of Amino Acids in Relation to their Cytosolic Levels in Barley Leaves 1. Plant Physiol. 1992, 99, 996–1004.
- Hayashi, H.; Chino, M. Collection of Pure Phloem Sap from Wheat and its Chemical Composition. Plant Cell Physiol. 1986, 27, 1387–1393.
- Hall, S.M.; Baker, D.A. The chemical composition of Ricinus phloem exudate. Planta 1972, 106, 131–140.
- Oner-Sieben, S.; Lohaus, G. Apoplastic and symplastic phloem loading in Quercus robur and Fraxinus excelsior. J. Exp. Bot. 2014, 65, 1905–1916.
- Fink, D.; Dobbelstein, E.; Barbian, A.; Lohaus, G. Ratio of sugar concentrations in the phloem sap and the cytosol of mesophyll cells in different tree species as an indicator of the phloem loading mechanism. Planta 2018, 248, 661–673.
- Pate, J.; Shedley, E.; Arthur, D.; Adams, M. Spatial and temporal variations in phloem sap composition of plantation-grown Eucalyptus globulus. Oecologia 1998, 117, 312–322.
- Killiny, N. Metabolite signature of the phloem sap of fourteen citrus varieties with different degrees of tolerance to Candidatus liberibacter asiaticus. Physiol. Mol. Plant Pathol. 2017, 97, 20–29.
- Heineke, D.; Sonnewald, U.; Büssis, D.; Günter, G.; Leidreiter, K.; Wilke, I.; Raschke, K.; Willmitzer, L.; Heldt, H.W. Apoplastic Expression of Yeast-Derived Invertase in Potato: Effects on Photosynthesis, Leaf Solute Composition, Water Relations, and Tuber Composition. Plant Physiol. 1992, 100, 301–308.
- Winzer, T.; Lohaus, G.; Heldt, H.-W. Influence of phloem transport, N-fertilization and ion accumulation on sucrose storage in the taproots of fodder beet and sugar beet. J. Exp. Bot. 1996, 47, 863–870.
- Hijaz, F.; Killiny, N. Collection and chemical composition of phloem sap from Citrus sinensis L. Osbeck (sweet orange). PLoS ONE 2014, 9, e101830.
- Yesbergenova-Cuny, Z.; Dinant, S.; Martin-Magniette, M.-L.; Quilleré, I.; Armengaud, P.; Monfalet, P.; Lea, P.J.; Hirel, B. Genetic variability of the phloem sap metabolite content of maize (Zea mays L.) during the kernel-filling period. Plant Sci. 2016, 252, 347–357.
- Stallmann, J.; Schweiger, R. Effects of Arbuscular Mycorrhiza on Primary Metabolites in Phloem Exudates of Plantago major and Poa annua and on a Generalist Aphid. Int. J. Mol. Sci. 2021, 22, 13086.
- Tedesco, S.; Erban, A.; Gupta, S.; Kopka, J.; Fevereiro, P.; Kragler, F.; Pina, A. The Impact of Metabolic Scion-Rootstock Interactions in Different Grapevine Tissues and Phloem Exudates. Metabolites 2021, 11, 349.
- Chardon, F.; De Marco, F.; Marmagne, A.; Le Hir, R.; Vilaine, F.; Bellini, C.; Dinant, S. Natural variation in the long-distance transport of nutrients and photoassimilates in response to N availability. J. Plant Physiol. 2022, 273, 153707.
- Fiehn, O. Metabolic networks of Cucurbita maxima phloem. Phytochemistry 2003, 62, 875–886.
- Sun, J.; Li, Q.; Xu, H.; Zhang, W. Analysis of Metabolomic Changes in Xylem and Phloem Sap of Cucumber under Phosphorus Stresses. Metabolites 2022, 12, 361.
- Gallinger, J.; Gross, J. Phloem Metabolites of Prunus sp. Rather than Infection with Candidatus phytoplasma prunorum Influence Feeding Behavior of Cacopsylla pruni Nymphs. J. Chem. Ecol. 2020, 46, 756–770.
- Peuke, A. Correlations in concentrations, xylem and phloem flows, and partitioning of elements and ions in intact plants. A summary and statistical re-evaluation of modelling experiments in Ricinus communis. J. Exp. Bot. 2010, 61, 635–655.
- Palmer, L.J.; Dias, D.A.; Boughton, B.; Roessner, U.; Graham, R.D.; Stangoulis, J.R. Metabolite profiling of wheat (Triticum aestivum L.) phloem exudate. Plant Methods 2014, 10, 27.
- Xie, H.; Shi, F.; Li, J.; Yu, M.; Yang, X.; Li, Y.; Fan, J. The Reciprocal Effect of Elevated CO2 and Drought on Wheat-Aphid Interaction System. Front. Plant Sci. 2022, 13, 853220.
- Turgeon, R.; Wolf, S. Phloem transport: Cellular pathways and molecular trafficking. Annu. Rev. Plant Biol. 2009, 60, 207–221.
- Liu, D.D.; Chao, W.M.; Turgeon, R. Transport of sucrose, not hexose, in the phloem. J. Exp. Bot. 2012, 63, 4315–4320.
- Guelette, B.S.; Benning, U.F.; Hoffmann-Benning, S. Identification of lipids and lipid-binding proteins in phloem exudates from Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 3603–3616.
- Valle, E.M.; Boggio, S.B.; Heldt, H.W. Free Amino Acid Composition of Phloem Sap and Growing Fruit of Lycopersicon esculentum. Plant Cell Physiol. 1998, 39, 458–461.
- Schneider, S.; GessLer, A.; Weber, P.; Sengbusch, D.; Hanemann, U.; Rennenberg, H. Soluble N compounds in trees exposed to high loads of N: A comparison of spruce (Picea abies) and beech (Fagus sylvatica) grown under field conditions. New Phytol. 1996, 134, 103–114.
- Hocking, P.J. The Composition of Phloem Exudate and Xylem Sap from Tree Tobacco (Nicotiana glauca Grah.). Ann. Bot. 1980, 45, 633–643.
- Lohaus, G.; Büker, M.; Hußmann, M.; Soave, C.; Heldt, H.-W. Transport of amino acids with special emphasis on the synthesis and transport of asparagine in the Illinois Low Protein and Illinois High Protein strains of maize. Planta 1998, 205, 181–188.
- Sandström, J.; Pettersson, J. Amino acid composition of phloem sap and the relation to intraspecific variation in pea aphid (Acyrthosiphon pisum) performance. J. Insect Physiol. 1994, 40, 947–955.
- Corbesier, L.; Havelange, A.; Lejeune, P.; Bernier, G.; Périlleux, C. N content of phloem and xylem exudates during the transition to flowering in Sinapis alba and Arabidopsis thaliana. Plant Cell Environ. 2001, 24, 367–375.
- Valim, M.F.; Killiny, N. Occurrence of free fatty acids in the phloem sap of different citrus varieties. Plant Signal. Behav. 2017, 12, e1327497.
- Hoffmann-Benning, S. Collection and Analysis of Phloem Lipids. Methods Mol. Biol. 2021, 2295, 351–361.
- Gaufichon, L.; Reisdorf-Cren, M.; Rothstein, S.; Chardon, F.; Suzuki, A. Biological functions of asparagine synthetase in plants. Plant Sci. 2010, 179, 141–153.
- Gaufichon, L.; Masclaux-Daubresse, C.; Tcherkez, G.; Reisdorf-Cren, M.; Sakakibara, Y.; Hase, T.; Clement, G.; Avice, J.C.; Grandjean, O.; Marmagne, A.; et al. Arabidopsis thaliana ASN2 encoding asparagine synthetase is involved in the control of nitrogen assimilation and export during vegetative growth. Plant Cell Environ. 2013, 36, 328–342.
- Kennecke, M.; Ziegler, H.; de Fekete, M.A. Enzyme activities in the sieve tube sap of Robinia pseudoacacia L. and of other tree species. Planta 1971, 98, 330–356.
- Knoblauch, M. Sieve Tubes in Action. Plant Cell 1998, 10, 35–50.
- Tilsner, J.; Kassner, N.; Struck, C.; Lohaus, G. Amino acid contents and transport in oilseed rape (Brassica napus L.) under different nitrogen conditions. Planta 2005, 221, 328–338.
- Girousse, C.; Bournoville, R.; Bonnemain, J.L. Water Deficit-Induced Changes in Concentrations in Proline and Some Other Amino Acids in the Phloem Sap of Alfalfa. Plant Physiol. 1996, 111, 109–113.
- Lothier, J.; Diab, H.; Cukier, C.; Limami, A.M.; Tcherkez, G. Metabolic Responses to Waterlogging Differ between Roots and Shoots and Reflect Phloem Transport Alteration in Medicago truncatula. Plants 2020, 9, 1373.
- van Beusichem, M.L.; Nelemans, O.A.; Hinnen, M.G.J. Nitrogen: Nitrogen cycling in plant species differing in shoot/root reduction of nitrate. J. Plant Nutr. 1987, 10, 1723–1731.
- Marschner, P.; Crowley, D.E.; Higashi, R.M. Root exudation and physiological status of a root-colonizing fluorescent pseudomonad in mycorrhizal and non-mycorrhizal pepper (Capsicum annuum L.). Plant Soil 1997, 189, 11–20.
- Teskey, R.O.; Saveyn, A.; Steppe, K.; McGuire, M.A. Origin, fate and significance of CO2 in tree stems. New Phytol. 2008, 177, 17–32.
- Trumbore, S.E.; Angert, A.; Kunert, N.; Muhr, J.; Chambers, J.Q. What’s the flux? Unraveling how CO2 fluxes from trees reflect underlying physiological processes. New Phytol. 2013, 197, 353–355.




