
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Zeinab Breijyeh | -- | 1780 | 2023-04-02 11:21:05 | | | |
| 2 | Peter Tang | Meta information modification | 1780 | 2023-04-03 04:24:58 | | |
Video Upload Options
Possible new ways for the development of novel classes of antibiotics are discussed here, for which there is no pre-existing resistance in human bacterial pathogens. By utilizing research and technology such as nanotechnology and computational methods (such as in silico and Fragment-based drug design (FBDD)), there has been an improvement in antimicrobial actions and selectivity with target sites. Moreover, there are antibiotic alternatives, such as antimicrobial peptides, essential oils, anti-Quorum sensing agents, darobactins, vitamin B6, bacteriophages, odilorhabdins, 18β-glycyrrhetinic acid, and cannabinoids. Additionally, drug repurposing (such as with ticagrelor, mitomycin C, auranofin, pentamidine, and zidovudine) and synthesis of novel antibacterial agents (including lactones, piperidinol, sugar-based bactericides, isoxazole, carbazole, pyrimidine, and pyrazole derivatives) represent novel approaches to treating infectious diseases. Nonetheless, prodrugs (e.g., siderophores) have shown to be an excellent platform to design a new generation of antimicrobial agents with better efficacy against multidrug-resistant bacteria.
1. Introduction
2. Antibiotic Classification
|
Antibiotics Family |
Mechanism of Action |
Antibiotics |
|---|---|---|
|
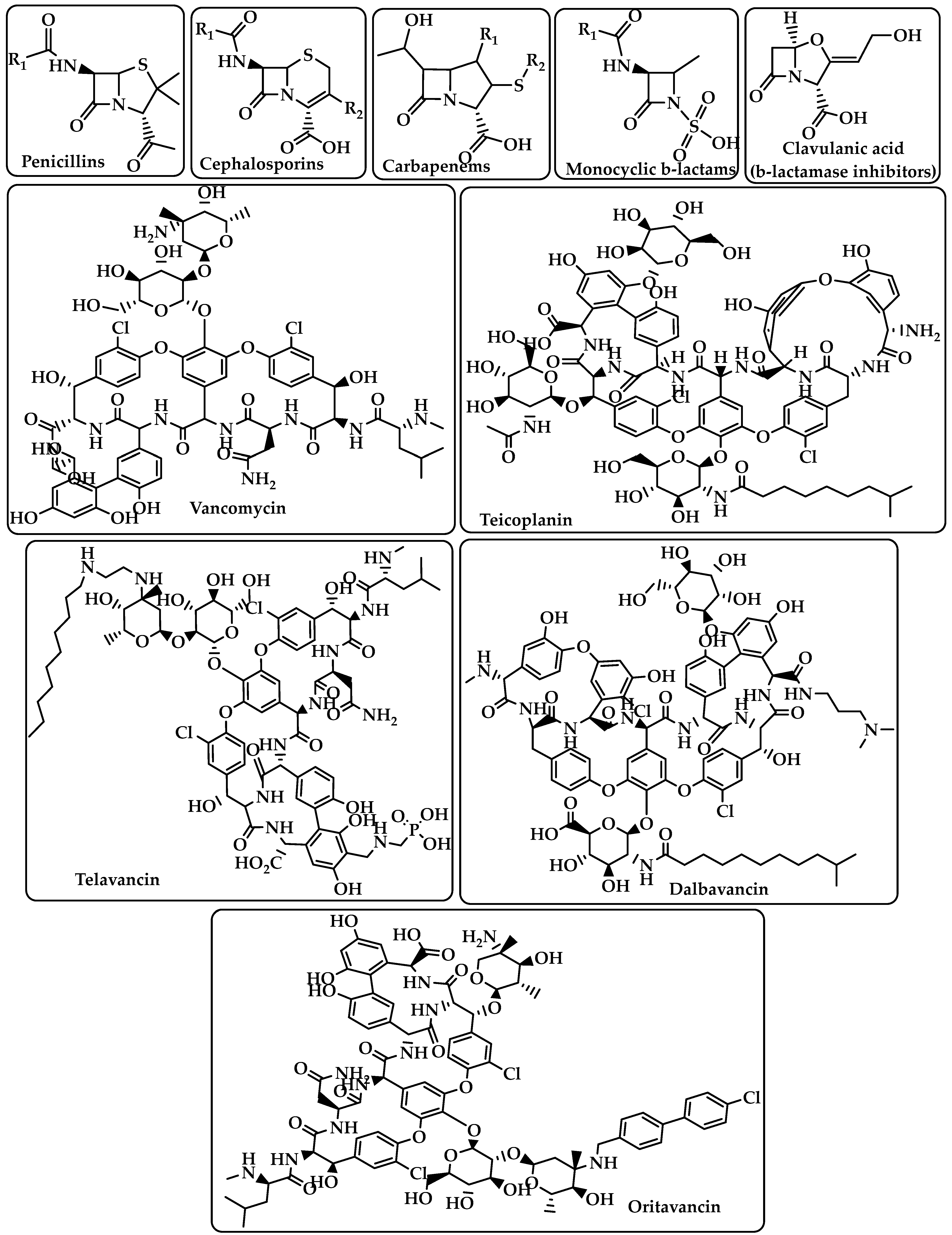
β-lactam |
Binds to the serine active site of penicillin-binding proteins (PBPs) or the allosteric site in PBP2a to inhibit bacterial cell wall peptidoglycan transpeptidation [14][15]. |
Penicillins Cephalosporins Carbapenems Monocyclic β-lactams β-lactamase inhibitors (e.g., clavulanic acid) (Figure 1) |
|
Glycopeptides |
Interacts with the membrane-bound lipid II precursor of peptidogly and can prevent peptidoglycan from being incorporated into an essential structural cell wall component [16]. |
Vancomycin Teicoplanin Telavancin Dalbavancin Oritavancin (Figure 1) |
|
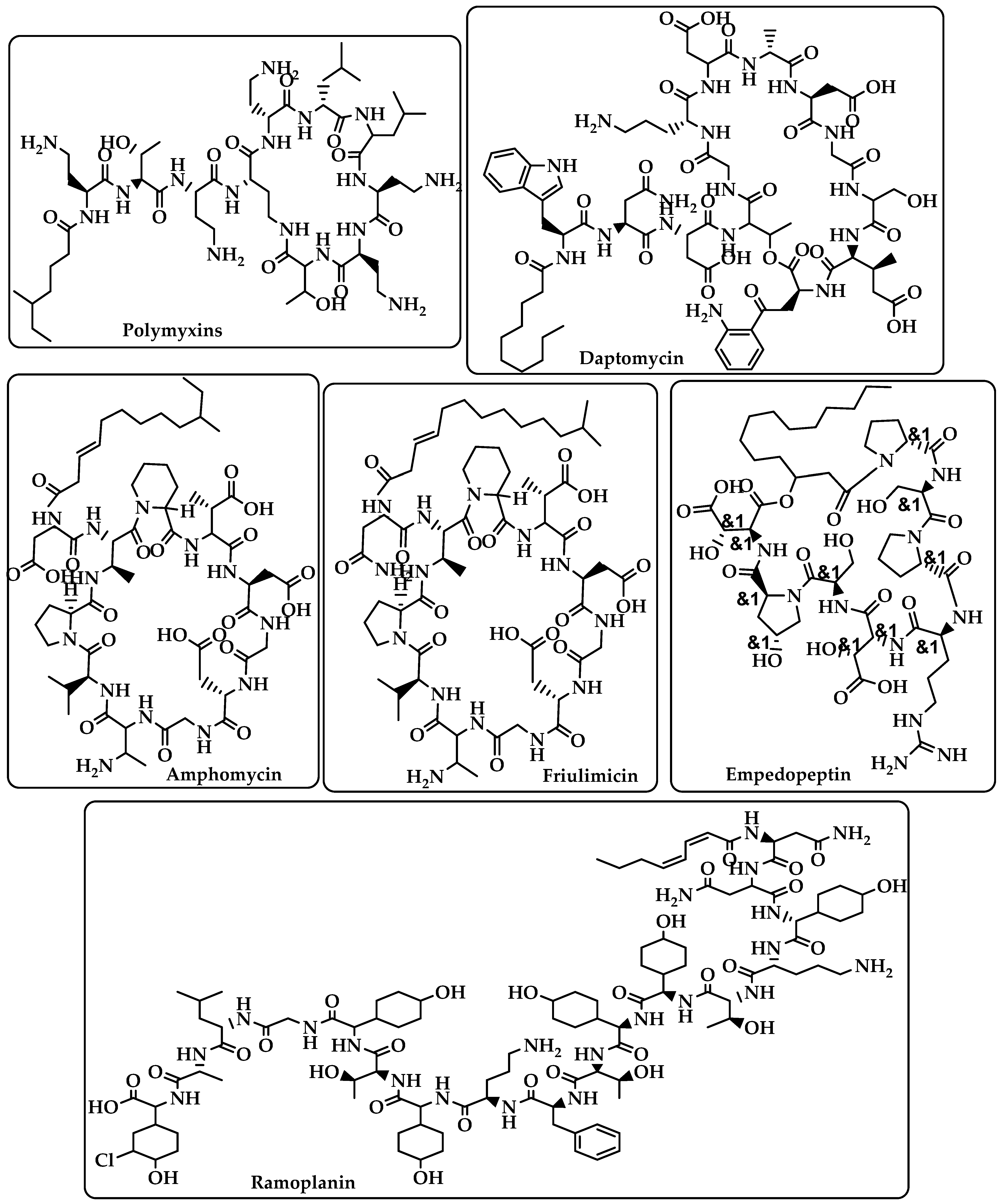
Lipopeptide |
Carries out their action by causing Gram-positive bacteria’s cell membrane integrity to be compromised, which results in cell death [17][18]. |
Polymyxins Daptomycin Amphomycin Friulimicin Ramoplanin Empedopeptin (Figure 2) |
|
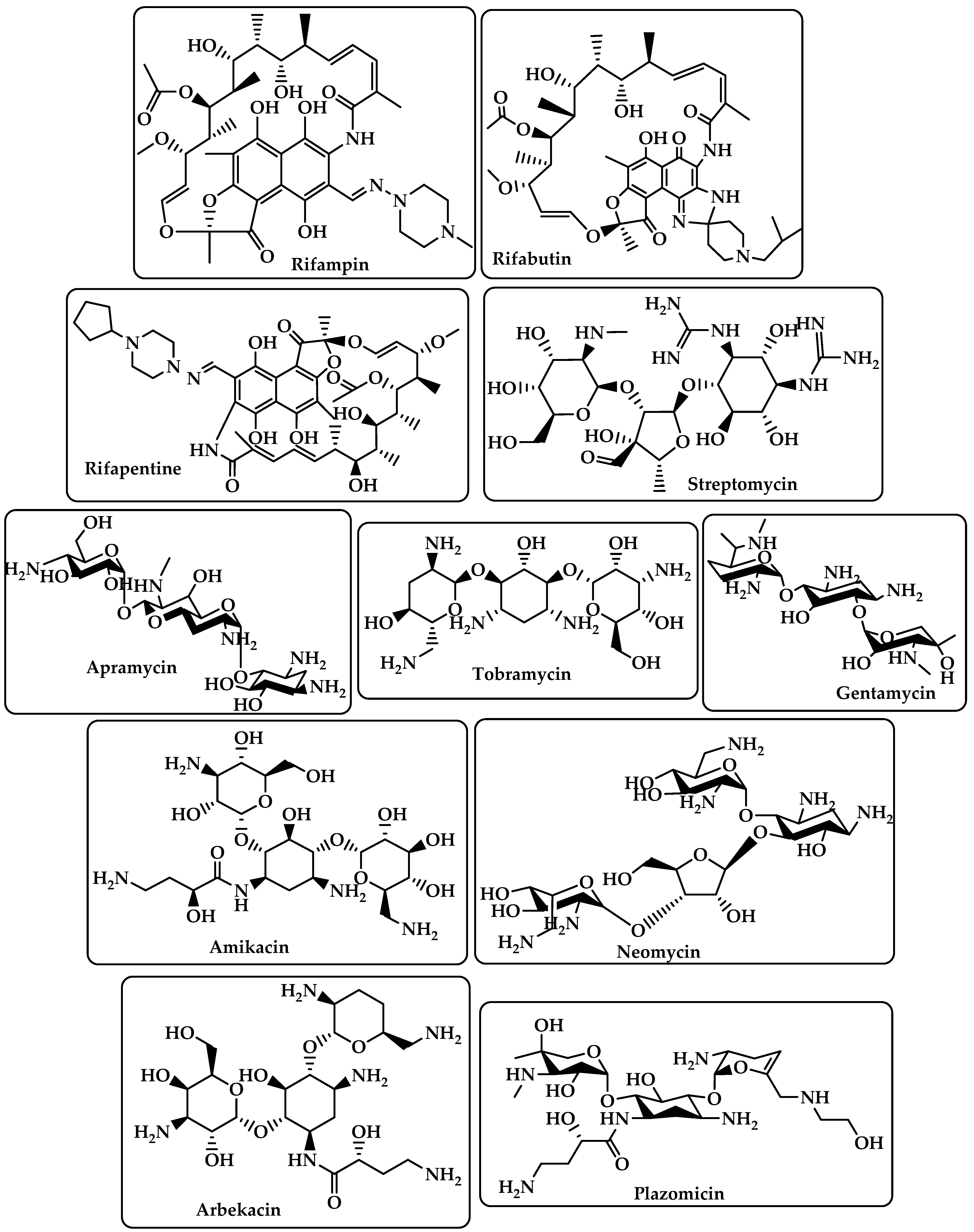
Rifamycins |
RNA polymerase (RNAP) inhibitors are used to treat tuberculosis (TB) [19]. |
Rifampin Rifabutin Rifapentine (Figure 3) |
|
Aminoglycoside |
By attaching to the 30S ribosome’s A-site on the 16S ribosomal RNA, they inhibit protein synthesis [20]. |
Streptomycin Apramycin Tobramycin Gentamcin Amikacin Neomycin Arbekacin Plazomicin (Figure 3) |
|
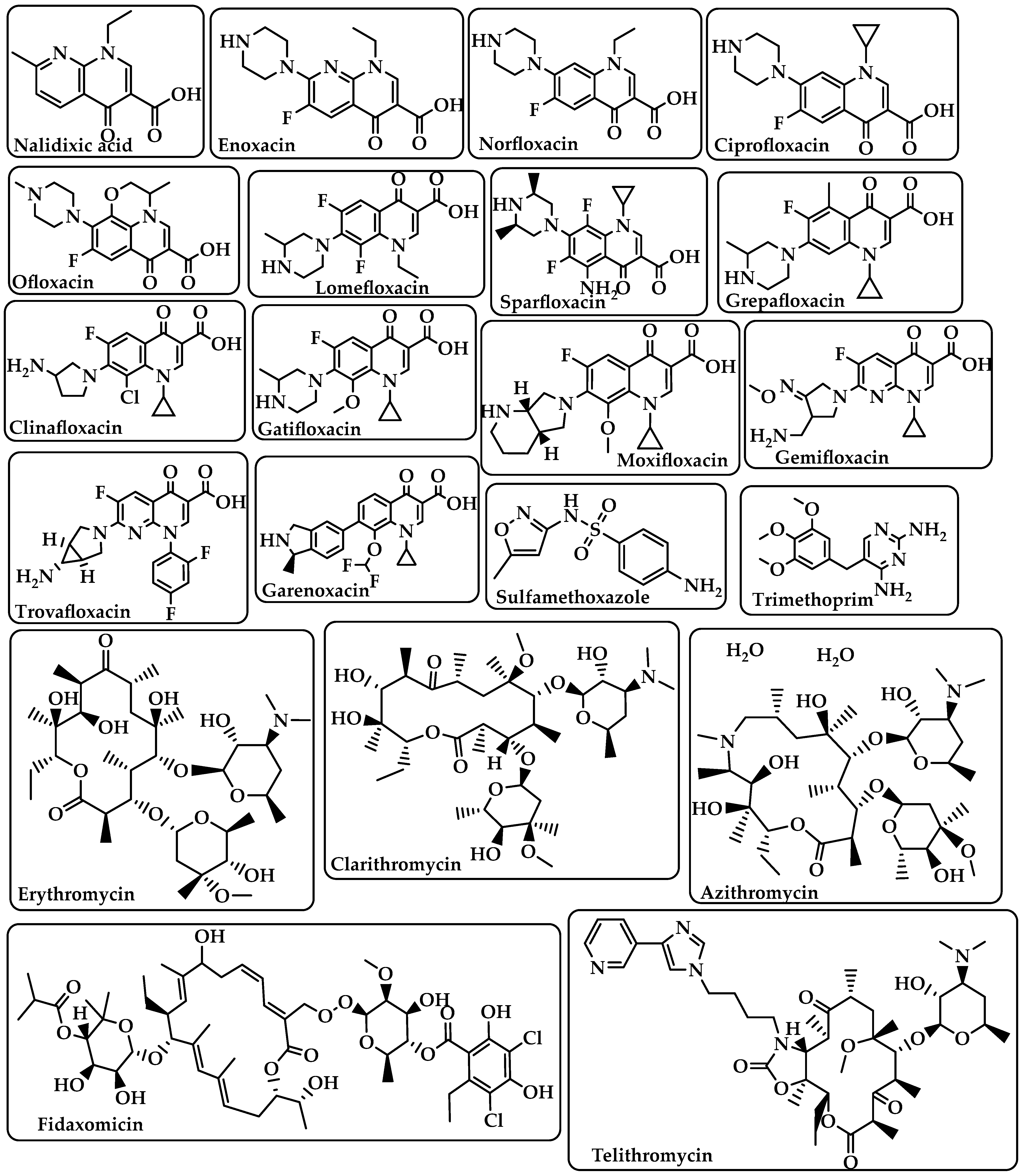
Fluoroquinolones |
Target DNA gyrase, topoisomerase IV, and topoisomerase type II to prevent bacteria from synthesizing DNA [21]. |
Nalidixic acid Enoxacin Norfloxacin Ciprofloxacin Ofloxacin Lomefloxacin Sparfloxacin Grepafloxacin Clinafloxacin Gatifloxacin Moxifloxacin Gemifloxacin Trovafloxacin Garenoxacin (Figure 4) |
|
Sulfonamides–Trimethoprim |
Sulfonamides interfere with the activity of the dihydropteroate synthase enzyme by competing with p-aminobenzoic acid (PABA) in the process of dihydrofolate production.The dihydrofolate reductase enzyme is inhibited by trimethoprim because it competes directly with it [22]. |
Sulfamethoxazole Trimethoprim (Figure 4) |
|
Macrolides |
Target the nascent peptide exit tunnel (NPET) of the bacterial 50S ribosomal subunit, which is responsible for the release of newly synthesized protein from the ribosome, ultimately preventing protein synthesis [23][24]. |
Erythromycin Clarithromycin Azithromycin Fidaxomicin Telithromycin (Figure 4) |
|
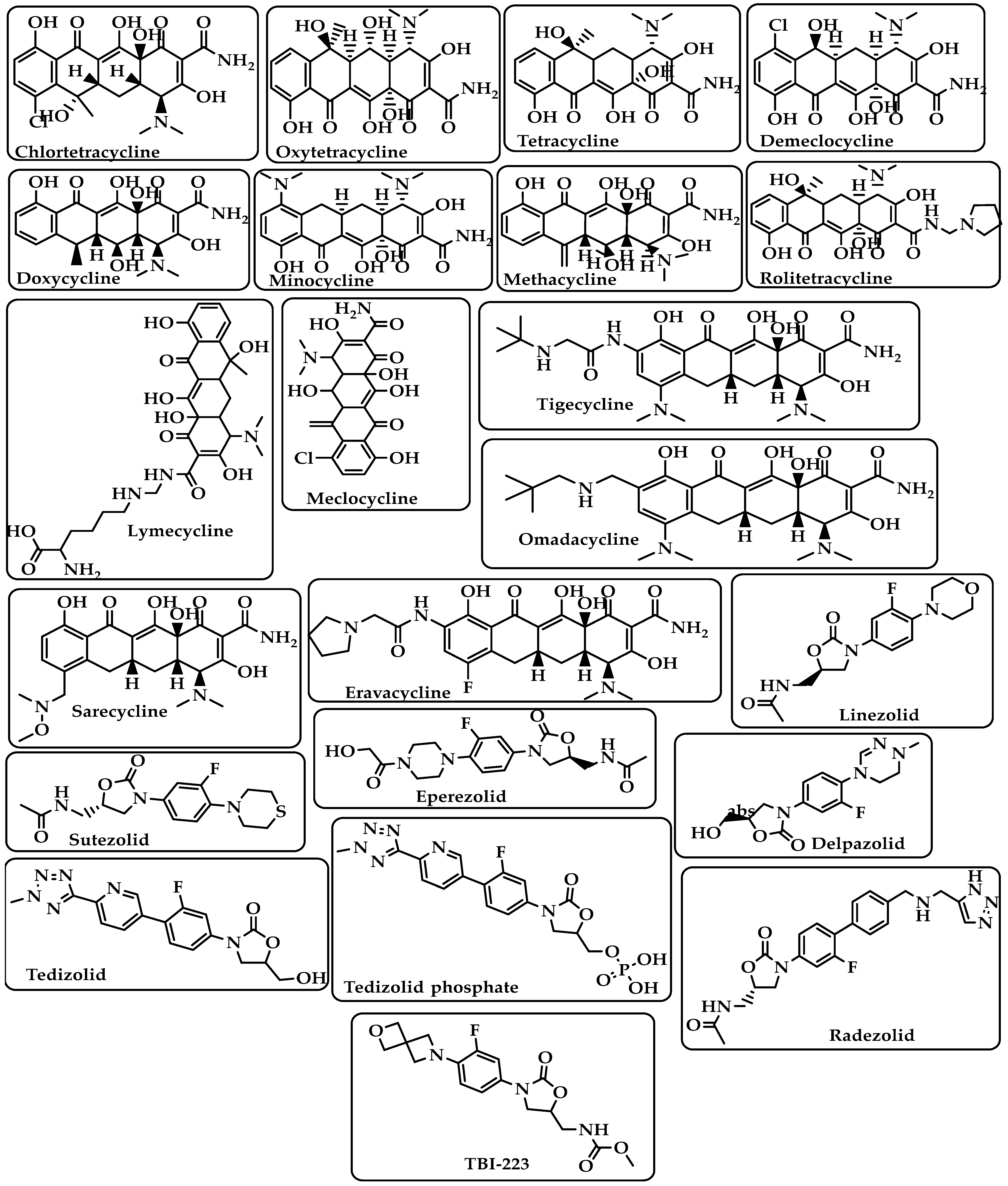
Tetracyclines |
Bind to the small subunit’s decoding site and prevent bacterial protein synthesis [25][26]. |
Chlortetracycline Oxytetracycline Tetracycline Demeclocycline Doxycycline Minocycline Lymecycline Meclocycline Methacycline RolitetracyclineTigecycline Omadacycline Sarecycline Eravacycline (Figure 5) |
|
Oxazolidinones |
Block the translation sequence by interacting with the 50S subunit (A-site pocket) at the peptidyl transferase center (PTC) to inhibit protein synthesis [27]. |
Linezolid Sutezolid Eperezolid Delpazolid Tedizolid Tedizolid phosphate Radezolid TBI-223 (Figure 5) |
|
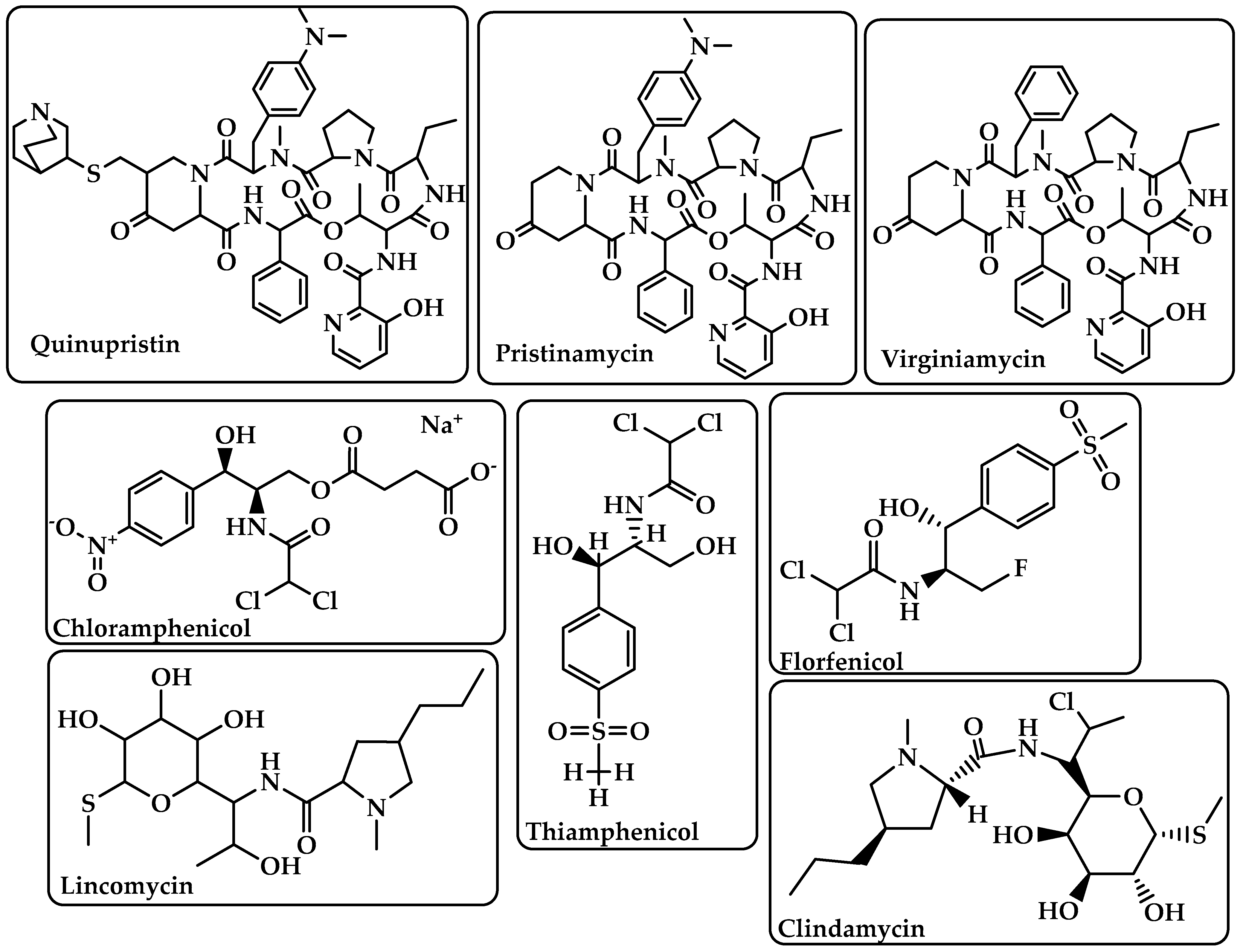
Streptogramins |
Inhibit protein synthesis during the elongation step by attaching to bacterial ribosomes [28]. The antibiotic has two unique structural groups (A and B) that cooperate to increase the affinity of group B in the nearby nascent peptide exit tunnel (NPET) when group A binds to the peptidyl transferase center (PTC) [29]. |
Quinupristin Pristinamycin Virginiamycin (Figure 6) |
|
Phenicoles |
Inhibit protein synthesis by binding to the 50S ribosomal subunit [30]. |
Chloramphenicol Thiamphenicol Florfenicol (Figure 6) |
|
Lincosamides |
Activate amino acid monomers by aminoacyl-tRNA, chain initiation, elongation, and termination of the formed polypeptides on the ribosome, which disrupts bacterial growth and death. These are only a few of the many processes that can be affected to prevent microbial protein synthesis [31]. |
Lincomycin Clindamycin (Figure 6) |
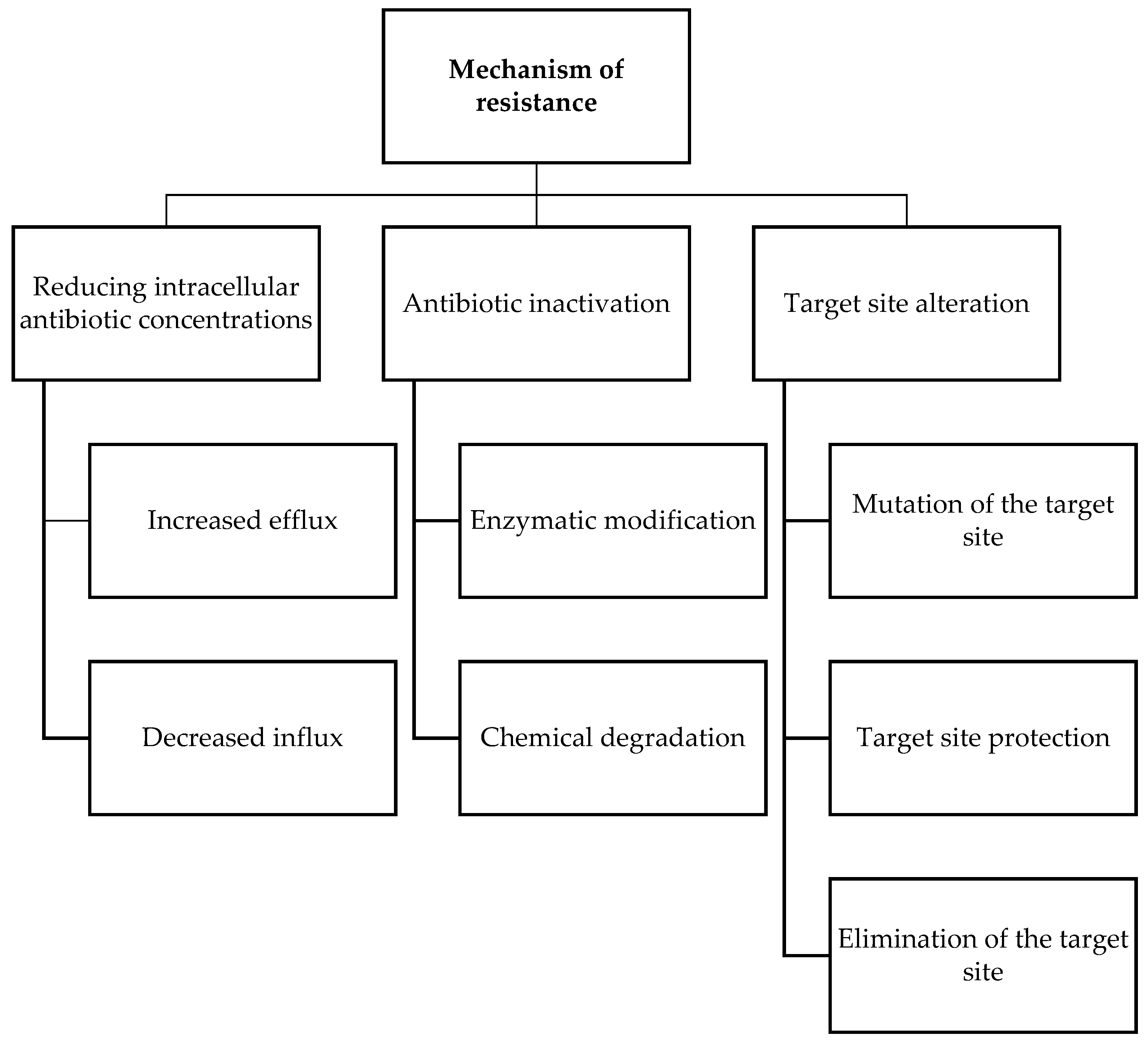
3. Antimicrobial Resistance
4. Antibiotic Use and Resistance in Agriculture Sector
References
- Adedeji, W.A. The Treasure Called Antibiotics. Ann. Ib. Postgrad. Med. 2016, 14, 56–57.
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340.
- Karaman, R.; Jubeh, B.; Breijyeh, Z. Resistance of Gram-Positive Bacteria to Current Antibacterial Agents and Overcoming Approaches. Molecules 2020, 25, 2888.
- Antimicrobial Resistance, C. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655.
- Asokan, G.V.; Ramadhan, T.; Ahmed, E.; Sanad, H. WHO Global Priority Pathogens List: A Bibliometric Analysis of Medline-PubMed for Knowledge Mobilization to Infection Prevention and Control Practices in Bahrain. Oman Med. J. 2019, 34, 184–193.
- WHO. Priority Pathogens List of Antibiotic-Resistant Bacteria for Which New Antibiotics are Urgently Needed; WHO: Geneva, Switzerland, 2017.
- Shrivastava, S.; Shrivastava, P.; Ramasamy, J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 2018, 32, 76.
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310.
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539.
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19.
- Qadri, H.; Shah, A.H.; Mir, M. Novel Strategies to Combat the Emerging Drug Resistance in Human Pathogenic Microbes. Curr. Drug Targets 2021, 22, 1424–1436.
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435.
- Calhoun, C.; Wermuth, H.R.; Hall, G.A. Antibiotics; StatPearls: Treasure Island, FL, USA, 2022.
- Bush, K.; Bradford, P.A. beta-Lactams and beta-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247.
- Eyler, R.F.; Shvets, K. Clinical Pharmacology of Antibiotics. Clin. J. Am. Soc. Nephrol. Cjasn 2019, 14, 1080–1090.
- Blaskovich, M.A.T.; Hansford, K.A.; Butler, M.S.; Jia, Z.; Mark, A.E.; Cooper, M.A. Developments in Glycopeptide Antibiotics. Acs Infect. Dis. 2018, 4, 715–735.
- Bionda, N.; Pitteloud, J.P.; Cudic, P. Cyclic lipodepsipeptides: A new class of antibacterial agents in the battle against resistant bacteria. Future Med. Chem. 2013, 5, 1311–1330.
- Schneider, T.; Muller, A.; Miess, H.; Gross, H. Cyclic lipopeptides as antibacterial agents—Potent antibiotic activity mediated by intriguing mode of actions. Int. J. Med. Microbiol. Ijmm 2014, 304, 37–43.
- Adams, R.A.; Leon, G.; Miller, N.M.; Reyes, S.P.; Thantrong, C.H.; Thokkadam, A.M.; Lemma, A.S.; Sivaloganathan, D.M.; Wan, X.; Brynildsen, M.P. Rifamycin antibiotics and the mechanisms of their failure. J. Antibiot. 2021, 74, 786–798.
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029.
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone antibiotics. Medchemcomm 2019, 10, 1719–1739.
- Kemnic, T.R.; Coleman, M. Trimethoprim Sulfamethoxazole; StatPearls: Treasure Island, FL, USA, 2022.
- Patel, P.H.; Hashmi, M.F. Macrolides; StatPearls: Treasure Island, FL, USA, 2022.
- Vazquez-Laslop, N.; Mankin, A.S. How Macrolide Antibiotics Work. Trends Biochem. Sci. 2018, 43, 668–684.
- Rusu, A.; Buta, E.L. The Development of Third-Generation Tetracycline Antibiotics and New Perspectives. Pharmaceutics 2021, 13, 2085.
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Donhofer, A.; Wilson, D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014, 395, 559–575.
- Foti, C.; Piperno, A.; Scala, A.; Giuffre, O. Oxazolidinone Antibiotics: Chemical, Biological and Analytical Aspects. Molecules 2021, 26, 4280.
- Bonfiglio, G.; Furneri, P.M. Novel streptogramin antibiotics. Expert Opin. Investig. Drugs 2001, 10, 185–198.
- Li, Q.; Pellegrino, J.; Lee, D.J.; Tran, A.A.; Chaires, H.A.; Wang, R.; Park, J.E.; Ji, K.; Chow, D.; Zhang, N.; et al. Synthetic group A streptogramin antibiotics that overcome Vat resistance. Nature 2020, 586, 145–150.
- Oong, G.C.; Tadi, P. Chloramphenicol; StatPearls: Treasure Island, FL, USA, 2022.
- Spizek, J.; Rezanka, T. Lincosamides: Chemical structure, biosynthesis, mechanism of action, resistance, and applications. Biochem Pharm. 2017, 133, 20–28.
- WHO. World Health Organization Ten Threats to Global Health in 2019; WHO: Geneva, Switzerland, 2019.
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. Aims Microbiol. 2018, 4, 482–501.
- Francine, P. Systems Biology: New Insight into Antibiotic Resistance. Microorganisms 2022, 10, 2362.
- Chancey, S.T.; Zahner, D.; Stephens, D.S. Acquired inducible antimicrobial resistance in Gram-positive bacteria. Future Microbiol. 2012, 7, 959–978.
- Mahon, C.R.; Lehman, D.C. Textbook of Diagnostic Microbiology—E-Book; Elsevier Health Sciences: USA, 2022.
- Sultan, I.; Rahman, S.; Jan, A.T.; Siddiqui, M.T.; Mondal, A.H.; Haq, Q.M.R. Antibiotics, Resistome and Resistance Mechanisms: A Bacterial Perspective. Front. Microbiol. 2018, 9, 2066.
- Ndagi, U.; Falaki, A.A.; Abdullahi, M.; Lawal, M.M.; Soliman, M.E. Antibiotic resistance: Bioinformatics-based understanding as a functional strategy for drug design. Rsc Adv. 2020, 10, 18451–18468.
- Kabra, R.; Chauhan, N.; Kumar, A.; Ingale, P.; Singh, S. Efflux pumps and antimicrobial resistance: Paradoxical components in systems genomics. Prog. Biophys. Mol. Biol. 2019, 141, 15–24.
- Ogawara, H. Comparison of Antibiotic Resistance Mechanisms in Antibiotic-Producing and Pathogenic Bacteria. Molecules 2019, 24, 3430.
- Spengler, G.; Kincses, A.; Gajdacs, M.; Amaral, L. New Roads Leading to Old Destinations: Efflux Pumps as Targets to Reverse Multidrug Resistance in Bacteria. Molecules 2017, 22, 468.
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 481–511.
- Schaenzer, A.J.; Wright, G.D. Antibiotic Resistance by Enzymatic Modification of Antibiotic Targets. Trends Mol. Med. 2020, 26, 768–782.
- Wilson, D.N.; Hauryliuk, V.; Atkinson, G.C.; O’Neill, A.J. Target protection as a key antibiotic resistance mechanism. Nat. Rev. Microbiol. 2020, 18, 637–648.
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795.
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 47.
- Finley, R.L.; Collignon, P.; Larsson, D.G.; McEwen, S.A.; Li, X.Z.; Gaze, W.H.; Reid-Smith, R.; Timinouni, M.; Graham, D.W.; Topp, E. The scourge of antibiotic resistance: The important role of the environment. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2013, 57, 704–710.
- Guetiya Wadoum, R.E.; Zambou, N.F.; Anyangwe, F.F.; Njimou, J.R.; Coman, M.M.; Verdenelli, M.C.; Cecchini, C.; Silvi, S.; Orpianesi, C.; Cresci, A.; et al. Abusive use of antibiotics in poultry farming in Cameroon and the public health implications. Br. Poult. Sci. 2016, 57, 483–493.
- Baynes, R.E.; Dedonder, K.; Kissell, L.; Mzyk, D.; Marmulak, T.; Smith, G.; Tell, L.; Gehring, R.; Davis, J.; Riviere, J.E. Health concerns and management of select veterinary drug residues. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2016, 88, 112–122.
- Maron, D.F.; Smith, T.J.; Nachman, K.E. Restrictions on antimicrobial use in food animal production: An international regulatory and economic survey. Glob. Health 2013, 9, 48.
- Gillings, M.R. Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Front. Microbiol. 2013, 4, 4.
- Dolliver, H.; Gupta, S. Antibiotic losses in leaching and surface runoff from manure-amended agricultural land. J. Environ. Qual. 2008, 37, 1227–1237.
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. Mmbr 2010, 74, 417–433.
- Phillips, I.; Casewell, M.; Cox, T.; De Groot, B.; Friis, C.; Jones, R.; Nightingale, C.; Preston, R.; Waddell, J. Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J. Antimicrob. Chemother. 2004, 53, 28–52.
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733.
- Moudgil, P.; Bedi, J.S.; Moudgil, A.D.; Gill, J.P.S.; Aulakh, R.S. Emerging issue of antibiotic resistance from food producing animals in India: Perspective and legal framework. Food Rev. Int. 2017, 34, 447–462.
- Sharma, C.; Rokana, N.; Chandra, M.; Singh, B.P.; Gulhane, R.D.; Gill, J.P.S.; Ray, P.; Puniya, A.K.; Panwar, H. Antimicrobial Resistance: Its Surveillance, Impact, and Alternative Management Strategies in Dairy Animals. Front. Vet. Sci. 2017, 4, 237.
- Ying, G.G.; He, L.Y.; Ying, A.J.; Zhang, Q.Q.; Liu, Y.S.; Zhao, J.L. China Must Reduce Its Antibiotic Use. Environ. Sci. Technol. 2017, 51, 1072–1073.
- Marquardt, R.R.; Li, S. Antimicrobial resistance in livestock: Advances and alternatives to antibiotics. Anim. Front. Rev. Mag. Anim. Agric. 2018, 8, 30–37.
- Rahman, M.R.T.; Fliss, I.; Biron, E. Insights in the Development and Uses of Alternatives to Antibiotic Growth Promoters in Poultry and Swine Production. Antibiotics 2022, 11, 766.




