
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Fang Zhu | -- | 2836 | 2023-03-14 18:53:05 | | | |
| 2 | Lindsay Dong | -1 word(s) | 2835 | 2023-03-15 06:22:12 | | |
Video Upload Options
Carboxyl/cholinesterases (CCEs) represent a family of enzymes distributed in many organisms, including insects. Despite their relatively simple catalyzed hydrolysis reaction, CCEs facilitate insects’ adaptation to chemical signals and stressors from the environment through various trajectories, including developing pesticide resistance, facilitating the adaptation of insects to their host plants, and manipulating insect behaviors. The CCE-mediated mechanisms of pesticide resistance to organophosphate, carbamate, or pyrethroid pesticides comprise enhanced metabolism, the sequestration of pesticides to prevent them from reaching their target sites, or conformational changes in target sites to prevent pesticides from binding. In addition, CCEs aid in the adaptation to chemical signals through the olfactory system by degrading insect semiochemicals.
1. Introduction
2. CCE Classification and Structural Characteristics
2.1. Classification of Insect CCEs
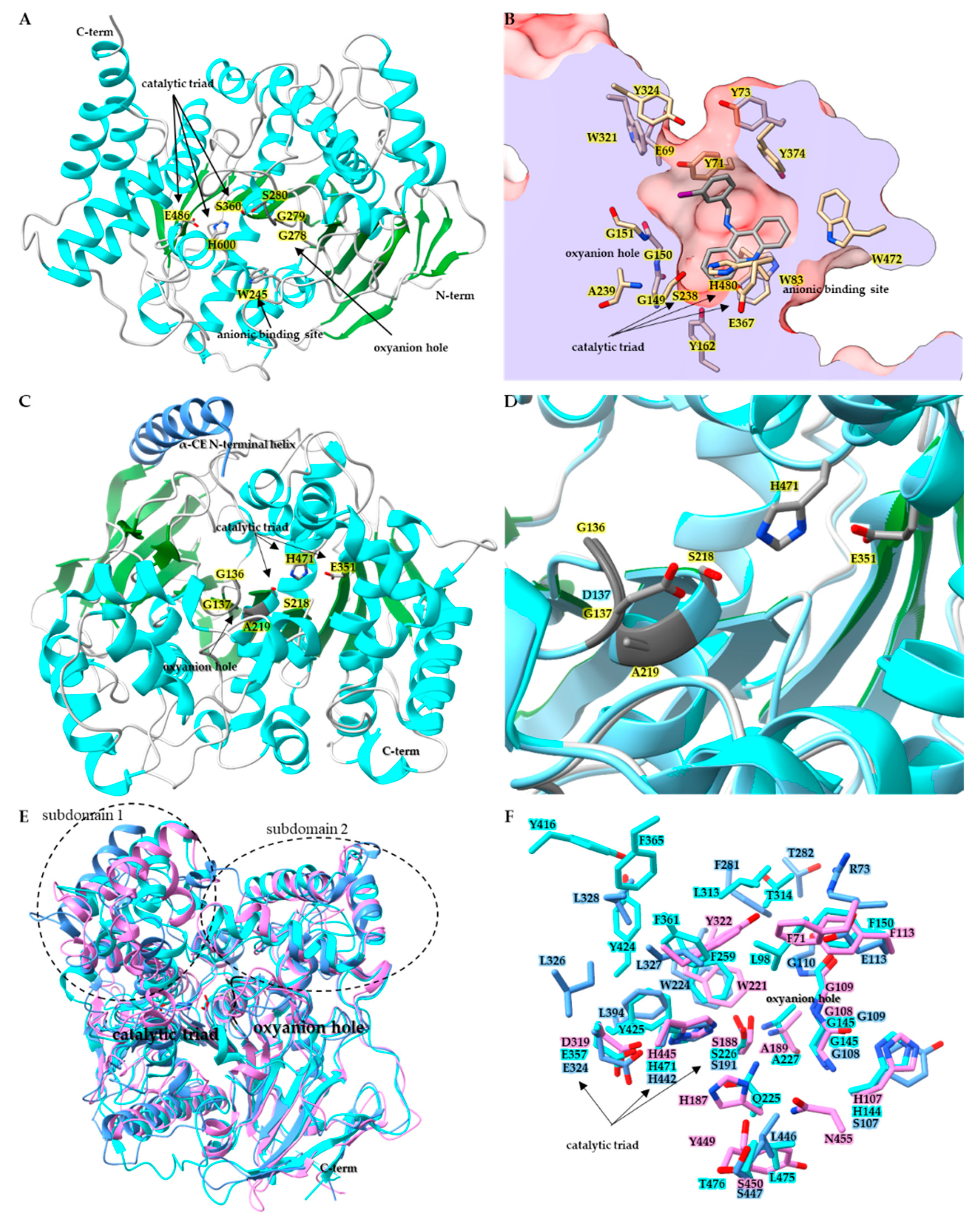
2.2. Structural Characteristics of Insect CCEs
3. Dynamic Rules of Insect CCEs in Chemical Adaptation
3.1. CCE-Mediated Insecticide Resistance
3.2. Metabolism of Plant Allelochemicals
3.3. Odorant Degradation in the Olfactory System
References
- Zhu, F.; Gujar, H.; Gordon, J.R.; Haynes, K.F.; Potter, M.F.; Palli, S.R. Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci. Rep. 2013, 3, 1456.
- Despres, L.; David, J.P.; Gallet, C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 2007, 22, 298–307.
- Koirala, B.K.S.; Moural, T.; Zhu, F. Functional and structural diversity of insect glutathione S-transferases in xenobiotic adaptation. Int. J. Biol. Sci. 2022, 18, 5713–5723.
- Brattsten, L.B. Biochemical defense mechansims in herbivores against plant allelochemicals. In Herbivores-Their Interaction with Secondary Plant Metabolites; Rosenthal, G.A., Janzen, D.H., Eds.; Academic Press: New York, NY, USA, 1979; pp. 199–270.
- Berenbaum, M.R.; Johnson, R.M. Xenobiotic detoxification pathways in honey bees. Curr. Opin. Insect. Sci. 2015, 10, 51–58.
- Zhu, F.; Cui, Y.; Walsh, D.B.; Lavine, L.C. Application of RNAi towards insecticide resistance management. In Short Views on Insect Biochemistry and Molecular Biology; Chandrasekar, R., Tyagi, B.K., Gui, Z., Reeck, G.R., Eds.; Academic Publisher: Manhattan, New York, NY, USA, 2014; Volume 2, pp. 595–619.
- Hamby, K.A.; Kwok, R.S.; Zalom, F.G.; Chiu, J.C. Integrating circadian activity and gene expression profiles to predict chronotoxicity of Drosophila suzukii response to insecticides. PLoS ONE 2013, 8, e68472.
- Amezian, D.; Nauen, R.; Le Goff, G. Transcriptional regulation of xenobiotic detoxification genes in insects—An overview. Pestic. Biochem. Physiol. 2021, 174, 104822.
- Montella, I.R.; Schama, R.; Valle, D. The classification of esterases: An important gene family involved in insecticide resistance—A review. Mem. Inst. Oswaldo Cruz 2012, 107, 437–449.
- Marshall, S.D.; Putterill, J.J.; Plummer, K.M.; Newcomb, R.D. The carboxylesterase gene family from Arabidopsis thaliana. J. Mol. Evol. 2003, 57, 487–500.
- Satoh, T.; Hosokawa, M. The mammalian carboxylesterases: From molecules to functions. Annu. Rev. Pharmacol. Toxicol. 1998, 38, 257–288.
- Bornscheuer, U.T. Microbial carboxyl esterases: Classification, properties and application in biocatalysis. FEMS Microbiol. Revs. 2002, 26, 73–81.
- Oakeshott, J.G.; Claudianos, C.; Russell, R.J.; Robin, G.C. Carboxyl/cholinesterases: A case study of the evolution of a successful multigene family. BioEssays 1999, 21, 1031–1042.
- Feng, X.; Li, M.; Liu, N. Carboxylesterase genes in pyrethroid resistant house flies, Musca domestica. Insect Biochem. Mol. Biol. 2018, 92, 30–39.
- Wei, H.; Tan, S.; Li, Z.; Li, J.; Moural, T.W.; Zhu, F.; Liu, X. Odorant degrading carboxylesterases modulate foraging and mating behaviors of Grapholita molesta. Chemosphere 2021, 270, 128647.
- Vogt, R.G. Molecular basis of pheromone detection in insects. In Comprehensive Molecular Insect Science; Gilbert, L.I., Iatro, K., Gill, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2005; Volume 3, pp. 753–804.
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253.
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391.
- Oakeshott, J.G.; Claudianos, C.; Campbell, P.M.; Newcomb, R.D.; Russell, R.J. Biochemical genetics and genomics of insect esterases. In Comprehensive Molecular Insect Science; Gilbert, L.I., Ed.; Elsevier: Amsterdam, The Netherlands, 2005; Volume 5, pp. 309–381.
- Oakeshott, J.G.; Johnson, R.M.; Berenbaum, M.R.; Ranson, H.; Cristino, A.S.; Claudianos, C. Metabolic enzymes associated with xenobiotic and chemosensory responses in Nasonia vitripennis. Insect Mol. Biol. 2010, 19 (Suppl. 1), 147–163.
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163.
- Ishida, Y.; Leal, W.S. Cloning of putative odorant-degrading enzyme and integumental esterase cDNAs from the wild silkmoth, Antheraea polyphemus. Insect Biochem. Mol. Biol. 2002, 32, 1775–1780.
- Klein, U. Sensillum-lymph proteins from antennal olfactory hairs of the moth Antheraea polyphemus (Saturniidae). Insect Biochem. 1987, 17, 1193–1204.
- Vogt, R.G.; Riddiford, L.M. Scale esterase: A pheromone-degrading enzyme from scales of silk moth Antheraea polyphemus. J. Chem. Ecol. 1986, 12, 469–482.
- Lu, F.G.; Fu, K.Y.; Li, Q.; Guo, W.C.; Ahmat, T.; Li, G.Q. Identification of carboxylesterase genes and their expression profiles in the Colorado potato beetle Leptinotarsa decemlineata treated with fipronil and cyhalothrin. Pestic. Biochem. Physiol. 2015, 122, 86–95.
- Yan, L.; Yang, P.; Jiang, F.; Cui, N.; Ma, E.; Qiao, C.; Cui, F. Transcriptomic and phylogenetic analysis of Culex pipiens quinquefasciatus for three detoxification gene families. BMC Genom. 2012, 13, 609.
- Gilbert, M.M.; Auld, V.J. Evolution of clams (cholinesterase-like adhesion molecules): Structure and function during development. Front. Biosci. 2005, 10, 2177–2192.
- Durand, N.; Chertemps, T.; Bozzolan, F.; Maïbèche, M. Expression and modulation of neuroligin and neurexin in the olfactory organ of the cotton leaf worm Spodoptera littoralis. Insect Sci. 2017, 24, 210–221.
- Zhu, K.Y.; Lee, S.H.; Clark, J.M. A point mutation of acetylcholinesterase associated with azinphosmethyl resistance and reduced fitness in Colorado potato beetle. Pestic. Biochem. Physiol. 1996, 55, 100–108.
- Baek, J.H.; Kim, J.I.; Lee, D.-W.; Chung, B.K.; Miyata, T.; Lee, S.H. Identification and characterization of ace1-type acetylcholinesterase likely associated with organophosphate resistance in Plutella xylostella. Pestic. Biochem. Physiol. 2005, 81, 164–175.
- Fournier, D. Mutations of acetylcholinesterase which confer insecticide resistance in insect populations. Chem. Biol. Interact. 2005, 157–158, 257–261.
- Lee, S.H.; Kim, Y.H.; Kwon, D.H.; Cha, D.J.; Kim, J.H. Mutation and duplication of arthropod acetylcholinesterase: Implications for pesticide resistance and tolerance. Pestic. Biochem. Physiol. 2015, 120, 118–124.
- Zhang, Y.; Yang, B.; Li, J.; Liu, M.; Liu, Z. Point mutations in acetylcholinesterase 1 associated with chlorpyrifos resistance in the brown planthopper, Nilaparvata lugens Stål. Insect Mol. Biol. 2017, 26, 453–460.
- Gilbert, M.; Smith, J.; Roskams, A.J.; Auld, V.J. Neuroligin 3 is a vertebrate gliotactin expressed in the olfactory ensheathing glia, a growth-promoting class of macroglia. Glia 2001, 34, 151–164.
- de la Escalera, S.; Bockamp, E.O.; Moya, F.; Piovant, M.; Jiménez, F. Characterization and gene cloning of neurotactin, a Drosophila transmembrane protein related to cholinesterases. EMBO J. 1990, 9, 3593–3601.
- Nardini, M.; Dijkstra, B.W. α/β Hydrolase fold enzymes: The family keeps growing. Curr. Opin. Struct. Biol. 1999, 9, 732–737.
- Wogulis, M.; Wheelock, C.E.; Kamita, S.G.; Hinton, A.C.; Whetstone, P.A.; Hammock, B.D.; Wilson, D.K. Structural studies of a potent insect maturation inhibitor bound to the juvenile hormone esterase of Manduca sexta. Biochem. J. 2006, 45, 4045–4057.
- Jackson, C.J.; Liu, J.W.; Carr, P.D.; Younus, F.; Coppin, C.; Meirelles, T.; Lethier, M.; Pandey, G.; Ollis, D.L.; Russell, R.J.; et al. Structure and function of an insect α-carboxylesterase (αEsterase7) associated with insecticide resistance. Proc. Natl. Acad. Sci. USA 2013, 110, 10177–10182.
- Harel, M.; Kryger, G.; Rosenberry, T.L.; Mallender, W.D.; Lewis, T.; Fletcher, R.J.; Guss, J.M.; Silman, I.; Sussman, J.L. Three-dimensional structures of Drosophila melanogaster acetylcholinesterase and of its complexes with two potent inhibitors. Preotein Sci. 2000, 9, 1063–1072.
- Hopkins, D.H.; Fraser, N.J.; Mabbitt, P.D.; Carr, P.D.; Oakeshott, J.G.; Jackson, C.J. Structure of an insecticide sequestering carboxylesterase from the disease vector Culex quinquefasciatus: What makes an enzyme a good insecticide sponge? Biochem. J. 2017, 56, 5512–5525.
- Kumar, K.; Mhetre, A.; Ratnaparkhi, G.S.; Kamat, S.S. A superfamily-wide activity atlas of Serine hydrolases in Drosophila melanogaster. Biochemistry 2021, 60, 1312–1324.
- Han, Q.; Wong, D.M.; Robinson, H.; Ding, H.; Lam, P.C.H.; Totrov, M.M.; Carlier, P.R.; Li, J. Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector Anopheles gambiae. Insect Sci. 2018, 25, 721–724.
- Cheung, J.; Mahmood, A.; Kalathur, R.; Liu, L.; Carlier, P.R. Structure of the G119S mutant acetylcholinesterase of the malaria vector Anopheles gambiae reveals basis of insecticide resistance. Structure 2018, 26, 130–136.e2.
- Younus, F.; Fraser, N.J.; Coppin, C.W.; Liu, J.-W.; Correy, G.J.; Chertemps, T.; Pandey, G.; Maïbèche, M.; Jackson, C.J.; Oakeshott, J.G. Molecular basis for the behavioral effects of the odorant degrading enzyme Esterase 6 in Drosophila. Sci. Rep. 2017, 7, 46188.
- Newcomb, R.D.; Campbell, P.M.; Ollis, D.L.; Cheah, E.; Russell, R.J.; Oakeshott, J.G. A single amino acid substitution converts a carboxylesterase to an organophosphorus hydrolase and confers insecticide resistance on a blowfly. Proc. Natl. Acad. Sci. USA 1997, 94, 7464–7468.
- Comoletti, D.; Trobiani, L.; Chatonnet, A.; Bourne, Y.; Marchot, P. Comparative mapping of selected structural determinants on the extracellular domains of cholinesterase-like cell-adhesion molecules. Neuropharmacology 2021, 184, 108381.
- Moural, T.W.; White, D.S.; Choy, C.J.; Kang, C.; Berkman, C.E. Crystal structure of phosphoserine blaC from mycobacterium tuberculosis inactivated by bis(benzoyl) phosphate. Int. J. Mol. Sci. 2019, 20, 3247.
- Ranson, H.; Claudianos, C.; Ortelli, F.; Abgrall, C.; Hemingway, J.; Sharakhova, M.V.; Unger, M.F.; Collins, F.H.; Feyereisen, R. Evolution of supergene families associated with insecticide resistance. N. Y. Sci. J. 2002, 298, 179–181.
- Soderlund, D.M. Molecular mechanisms of pyrethroid insecticide neurotoxicity: Recent advances. Arch. Toxicol. 2012, 86, 165–181.
- Feyereisen, R.; Dermauw, W.; Van Leeuwen, T. Genotype to phenotype, the molecular and physiological dimensions of resistance in arthropods. Pestic. Biochem. Physiol. 2015, 121, 61–77.
- Cui, F.; Li, M.X.; Chang, H.J.; Mao, Y.; Zhang, H.Y.; Lu, L.X.; Yan, S.G.; Lang, M.L.; Liu, L.; Qiao, C.L. Carboxylesterase-mediated insecticide resistance: Quantitative increase induces broader metabolic resistance than qualitative change. Pestic. Biochem. Physiol. 2015, 121, 88–96.
- Mutero, A.; Pralavorio, M.; Bride, J.M.; Fournier, D. Resistance-associated point mutations in insecticide-insensitive acetylcholinesterase. Proc. Natl. Acad. Sci. USA 1994, 91, 5922–5926.
- Weill, M.; Malcolm, C.; Chandre, F.; Mogensen, K.; Berthomieu, A.; Marquine, M.; Raymond, M. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol. Biol. 2004, 13, 1–7.
- Field, L.M.; Blackman, R.L.; Tyler-Smith, C.; Devonshire, A.L. Relationship between amount of esterase and gene copy number in insecticide-resistant Myzus persicae (Sulzer). Biochem. J. 1999, 339 (Pt 3), 737–742.
- Devonshire, A.L.; Moores, G.D. A carboxylesterase with broad substrate specificity causes organophosphorus, carbamate and pyrethroid resistance in peach-potato aphids (Myzus persicae). Pestic. Biochem. Physiol. 1982, 18, 235–246.
- Tang, B.; Dai, W.; Qi, L.; Du, S.; Zhang, C. Functional characterization of an α-esterase gene associated with malathion detoxification in Bradysia odoriphaga. J. Agric. Food Chem. 2020, 68, 6076–6083.
- Whyard, S.; Downe, A.E.; Walker, V.K. Characterization of a novel esterase conferring insecticide resistance in the mosquito Culex tarsalis. Arch. Insect Biochem. Physiol. 1995, 29, 329–342.
- Yang, X.; Margolies, D.C.; Zhu, K.Y.; Buschman, L.L. Host plant-induced changes in detoxification enzymes and susceptibility to pesticides in the twospotted spider mite (Acari: Tetranychidae). J. Econ. Entomol. 2001, 94, 381–387.
- Yang, Z.; Zhang, F.; He, Q.; He, G. Molecular dynamics of detoxification and toxin-tolerance genes in brown planthopper (Nilaparvata lugens Stål., Homoptera: Delphacidae) feeding on resistant rice plants. Arch. Insect Biochem. Physiol. 2005, 59, 59–66.
- Karuppaiah, V.; Srivastava, C.; Subramanian, S. Effect of host plants on insecticide susceptibility and detoxification enzymes activity in Spodoptera litura Fabricius (Noctuidae: Lepidoptera). Proc. Natl. Acad. Sci. India B-Biol. Sci. 2016, 86, 715–721.
- Liu, B.; Coy, M.; Wang, J.J.; Stelinski, L.L. The effect of host plant species on the detoxifying enzymes of the Asian citrus psyllid, Diaphorina citri (Hemiptera: Liviidae). Fla. Entomol. 2015, 98, 997–999.
- Xu, H.X.; Hong, Y.; Zhang, M.Z.; Wang, Y.L.; Liu, S.S.; Wang, X.W. Transcriptional responses of invasive and indigenous whiteflies to different host plants reveal their disparate capacity of adaptation. Sci. Rep. 2015, 5, 10774.
- Zhao, C.; Zhu, F.; Sun, Q.; Zhou, X. Editorial: Mechanisms and strategies of arthropod adaptation to the chemical environment. Front. Physiol. 2022, 13, 889757.
- Lu, K.; Song, Y.; Zeng, R. The role of cytochrome P450-mediated detoxification in insect adaptation to xenobiotics. Curr. Opin. Insect. Sci. 2021, 43, 103–107.
- Zhu, F.; Moural, T.W.; Nelson, D.R.; Palli, S.R. A specialist herbivore pest adaptation to xenobiotics through up-regulation of multiple Cytochrome P450s. Sci. Rep. 2016, 6, 20421.
- Scriber, J.M.; Lindroth, R.L.; Nitao, J.K. Toxic phenolic glycosides from populus: Physiological adaptations of the Western North American tiger awallowtail butterfly, Papilio rutulus (Lepidoptera: Papilionidae). Gt. Lakes Entomol. 1991, 24, 173–180.
- Lindroth, R.L.; Weisbrod, A.V. Genetic-variation in response of the Gypsy-moth to aspen phenolic glycosides. Biochem. Syst. Ecol. 1991, 19, 97–103.
- Hemming, J.D.C.; Lindroth, R.L. Effects of phenolic glycosides and protein on gypsy moth (Lepidoptera: Lymantriidae) and forest tent caterpillar (Lepidoptera: Lasiocampidae) performance and detoxication activities. Environ. Entomol. 2000, 29, 1108–1115.
- Cai, Q.N.; Han, Y.; Cao, Y.Z.; Hu, Y.; Zhao, X.; Bi, J.L. Detoxification of gramine by the cereal aphid Sitobion avenae. J. Chem. Ecol. 2009, 35, 320–325.
- Song, L.-M.; Jiang, X.; Wang, X.-M.; Li, J.-D.; Zhu, F.; Tu, X.-B.; Zhang, Z.-H.; Ban, L.-P. Male tarsi specific odorant-binding proteins in the diving beetle Cybister japonicus Sharp. Sci. Rep. 2016, 6, 31848.
- Homberg, U.; Christensen, T.A.; Hildebrand, J.G. Structure and function of the deutocerebrum in insects. Annu. Rev. Entomol. 1989, 34, 477–501.
- Wu, H.; Liu, Y.; Shi, X.; Zhang, X.; Ye, C.; Zhu, K.Y.; Zhu, F.; Zhang, J.; Ma, E. Transcriptome analysis of antennal cytochrome P450s and their transcriptional responses to plant and locust volatiles in Locusta migratoria. Int. J. Biol. Macromol. 2020, 149, 741–753.
- Younus, F.; Chertemps, T.; Pearce, S.L.; Pandey, G.; Bozzolan, F.; Coppin, C.W.; Russell, R.J.; Maibeche-Coisne, M.; Oakeshott, J.G. Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster. Insect Biochem. Mol. Biol. 2014, 53, 30–43.
- Steiner, C.; Chertemps, T.; Maïbèche, M. Diversity of biotransformation enzymes in insect antennae: Possible roles in odorant inactivation and xenobiotic processing. In Olfactory Concepts of Insect Control—Alternative to Insecticides; Picimbon, J.-F., Ed.; Springer International Publishing: Cham, Switzerland, 2019; Volume 2, pp. 115–145.
- Durand, N.; Carot-Sans, G.; Chertemps, T.; Bozzolan, F.; Party, V.; Renou, M.; Debernard, S.; Rosell, G.; Maïbèche-Coisne, M. Characterization of an antennal carboxylesterase from the pest moth Spodoptera littoralis degrading a host plant odorant. PLoS ONE 2010, 5, e15026.
- Durand, N.; Carot-Sans, G.; Bozzolan, F.; Rosell, G.; Siaussat, D.; Debernard, S.; Chertemps, T.; Maïbèche-Coisne, M. Degradation of pheromone and plant volatile components by a same odorant-degrading enzyme in the cotton leafworm, Spodoptera littoralis. PLoS ONE 2011, 6, 29147.




