1. Introduction
Microfluidic organ-on-chip (OoC) is an emerging technology comprising bioengineered human tissues housed within microfluidic devices. OoCs leverage microfabrication technologies to achieve spatio-temporal control over various cell microenvironmental factors at the cellular length scale (between 10–100 microns) (
Figure 1) and, therefore, can better mimic the physiological functions of various tissues
[1]. The miniaturised and multiplexable nature of OoCs makes them an attractive platform for disease modelling and drug testing, and interest is growing in OoC technologies for generating organotypic tissues, such as the subchondral bone tissues (covered in existing reviews)
[2][3]. While the generation of organotypic tissues can improve OoC relevance in OA models and drug screening, control of the tissue environment is essential to maintain native tissue function
[4]. Due to the complexity of the joint tissue, many joint OoCs use a deconstructed approach to modelling specific cell environments, allowing the systemic investigation of their role in OA. These strategies can potentially help improve the applicability of in vitro OoC models for disease modelling and drug testing. There has been significant progress in using microfluidics to control various microenvironmental factors relevant to different aspects of joint tissue physiology (
Figure 1) in the context of OA, which will be reviewed and discussed herein, along with bone and cartilage OoCs as most OA OoC models are built on these two models.
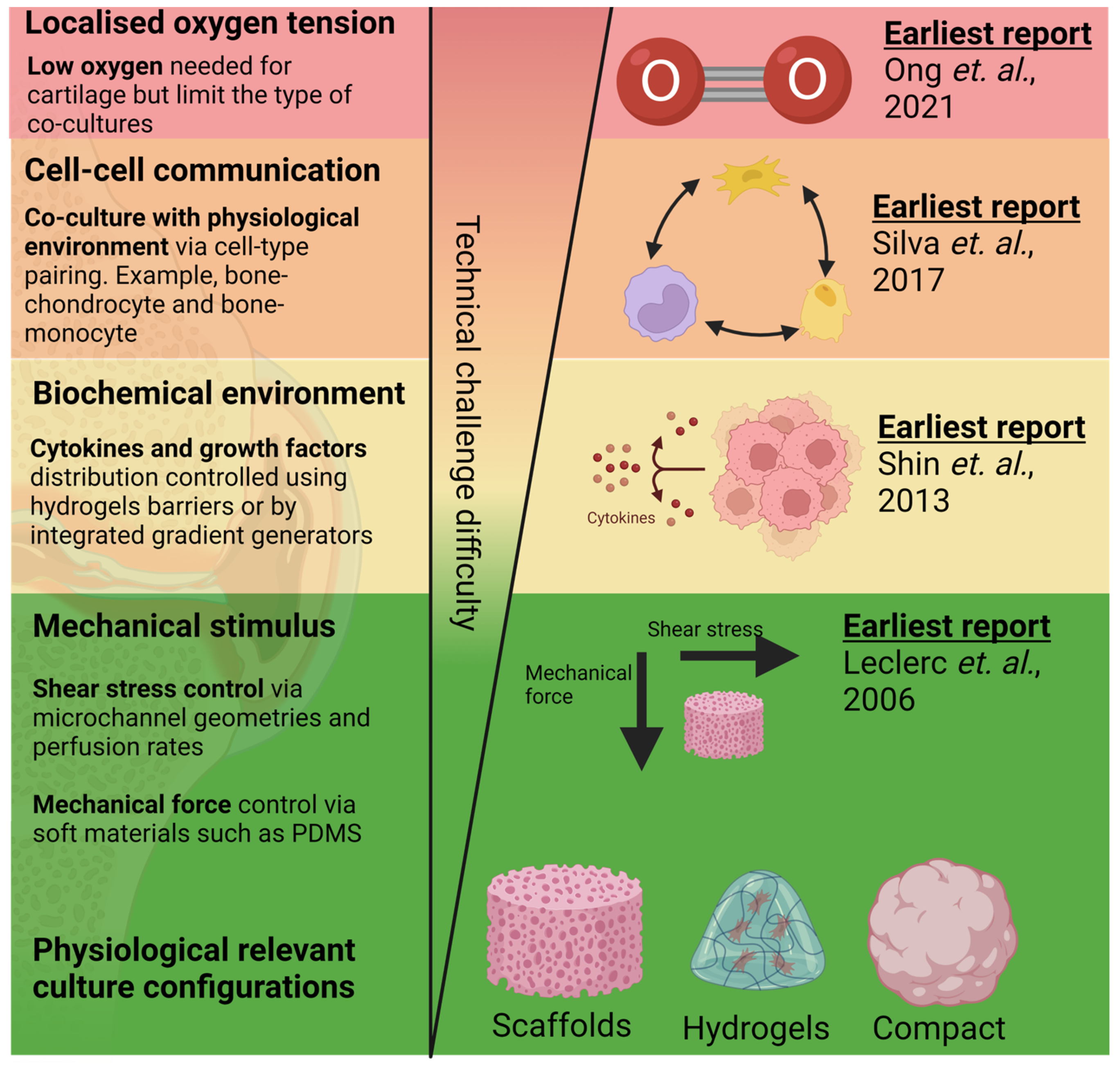
Figure 1. Reported work on mimicking aspects of OA tissue physiology in order of technical challenges in the design, fabrication, and operation of cartilage and bone microfluidic cultures. Techniques to incorporate 3D culture
[5] and mechanical
[6] and biochemical stimulus
[7] are among the earliest techniques developed and have been relatively matured. Alternatively, modelling of oxygen tension control
[8] and multi-cell co-cultures
[9] are comparatively new techniques and might require greater optimisation for their adoption into routine tissue culture workflow. Created with
BioRender.com.
2. Bioengineering 3D Bone and Cartilage Tissues in OoCs
With growing evidence of the importance of 3D cultures in providing physiologically relevant tissue response
[10], most of the culture for both bone and cartilage uses various strategies to achieve 3D cultures in microfluidic OoC devices. Techniques modelling 3D tissue structure in OoC are relatively well developed (
Figure 2), with the earliest report of OoC design of an OA model noted in 2006
[5]. Many reported 3D cartilage and bone OoC leverages 3D tissue culture techniques developed for other organs. For example, Rosser’s work demonstrated the significance of 3D microfluidic culture where chondrocytes exhibited a higher differentiated cartilage phenotype and osteoarthritis mimicking response to inflammation and treatments compared to 2D monolayers
[11]. Similarly, Choudhary et al. also showed that 3D microfluidic ex vivo culture of primary osteocytes could help retain mature osteocytic phenotypes (SOST, FGF23) better than 2D monolayers
[12], which plays a crucial role in OA disease progression
[13].
For cartilage models, hydrogels were predominantly in OoCs
[11][14][15][16][17][18][19][20]. This approach is common in device cultures for the hydrogel to be capable of mimicking the ECM of cartilage. Since cartilage ECM typically contains a mixture of type I and type II collagen, a shift in the collagen content within the ECM could change the function of the chondrocyte
[4]. Incorporating cell-laden hydrogels into microfluidic devices can be achieved by either loading liquid-phase hydrogels into a device before setting or by placing crosslinked hydrogels into dedicated chambers.
Liquid-phase hydrogels are typically loaded through a straight channel with microstructures to guide the hydrogel flow. For example, Paggi et al. reported a microfluidic device with channels consisting of lined micropillar arrays, partitioning chondrocytes laden agarose within the microchannel, which were then perfused with culture media
[14]. This partitioning allowed 3D cultures of chondrocytes on one side of the microchannels while allowing perfusion on the adjacent chambers
[11][14]. This technique relied on the micropillar arrays at specific spatial intervals (typically around 20–30 µm) to hold hydrogels in the liquid phase via surface tension during the loading process
[7]. Therefore, this technique would require high-resolution microfabrication foundries. Changing channel geometry is a recent and alternative strategy exploiting hydrogel’s surface tension to enable hydrogel pinning within an open channel. For example, Hou et al. reported a PDMS microchannel device with a 100 µm high device channel positioned next to a microchannel with a higher ceiling
[21]. The change in microchannel ceiling functions similarly to the micropillars used by Roger Kamm’s team but alleviates users from the need for high-resolution fabrication techniques
[7]. As surface tension depends on the hydrophobicity of the device’s surface material, it is essential to consider the fabrication materials used. With 3D printing adopted increasingly in the fabrication of plastic-based microfluidic devices, the selection of plastic materials and surface modification techniques to modify surface hydrophobicity should be considered during the device design and fabrication process. A similar strategy is also observed for bone cultures
[22][23], where, for example, Ma et al. used cell-laden Matrigel to compartmentalise osteocytes in microfluidic devices
[22].
A different technique for capturing chondrocyte 3D tissue configuration is densely packing the cells into micro-compartments. Dense-packed chondrocyte pellets, commonly thought to mimic mesenchymal condensation
[22], showed an enhanced COL2A: COL1A ratio, which can determine the disease progression in OA. Yang et al. reported the generation of a microfluidic platform with arrays of concave microwells to allow seeded chondrocytes to sediment into the microwells and form 3D micro pellets
[24]. The microwell array provides a lower technical barrier for the user to consistently create 3D cell cultures at the microscale, compared with 3D cultures relying on hydrogels. Additionally, this strategy is amendable for various microscopy platforms for high throughput screening and data sampling processes. However, additional design modifications for the microwell-based devices are needed for the platform to be amendable to perfusion cultures. Given that bone tissue does not form dense packing, no work on compact bone cell cultures has been identified for OA models.
Incorporating scaffolds to create 3D tissue models for chondrocytes and bone are also a common strategy
[25][26][27]. The stiffness of the scaffold is tuneable to mimic cartilage tissue stiffness, which is essential for maintaining and supporting the differentiation of functional chondrocytes
[28]. However, the scaffold material and fabrication process are typically incompatible with standard soft lithography, as creating porous PDMS structures remains challenging. As such, design for scaffold-based microfluidic cultures would consider an additional step to include assembly of the scaffold in the device. The most straightforward method is to fabricate the device and scaffold independently, followed by sequential assembly of the scaffold by plasma bonding, clamping, or chemical sealants. Mekhileri et al. reported using 3D bioprinting to first print PEGT/PBT thermoplastics as a scaffold to house the chondrocytes laden within GelMa layers
[29]. With careful device design, these scaffolds can be integrated into microfluidic devices to subject the cultures to fluid shear stress. In another study, Sieber et al. incorporated ceramic-based scaffolds into PDMS devices to develop a microfluidic bone model
[25]. One potential advantage of this fabrication strategy is that it reduces the need for high-resolution fabrication typically needed to generate porous structures. With hydrogel loading, the microchambers and channels should be designed to prevent blockage of perfusion channels during gel loading (
Figure 2).
Since different aspects of tissue microenvironment can be achieved across the common culture modalities
[30], demonstration of OoC culture of chondrocyte cultures often uses a combination of hydrogel with either scaffolds or pellet cultures. For example, Rothbauer et al. cultured densely packed chondrocytes embedded in Matrigel to mimic cell–cell and cell–ECM interactions within the cartilage matrix
[31].
Figure 2. Key strategies implemented in OoC that are adaptable for mimicking joint physiologies for OA models. (
A) Integrated hydrogels or scaffolds can achieve 3D culture in OoC
[19][26]. (
B) Compressive forces are typically mimicked with an elastic material such as PDMS to allow the channel walls to be compressed using a pneumatic system
[14]. (
C) Through controlling the laminar stream, concentration generators
[32] can be integrated on-chip to control concentrations of cytokines and chemokines mimicking OA inflammation responses. (
D) Cell–cell interactions are typically mimicked through compartmentalised culture models connected with liquid microchannels
[31]. Created with
BioRender.com. All Figures are obtained under Creative Commons Attribution 3.0 Unported Licence.
3. Application of Mechanical and Fluid-Induced Shear Stress
With growing evidence of the role of physical activities as a potential treatment of OA, modelling mechanical signalling in microfluidic bone and tissue culture either induced by interstitial fluid motion or through forces subjected during joint loading could provide a suitable animal alternative to screen treatment efficacies for OA. With almost all microfluidic devices holding a volume of less than 500 μL, microfluidic cultures almost always relied on perfusion to facilitate the mass transport of nutrients during cell cultures, which generate fluid-induced shear stress. The shear stress can be controlled by manipulating channel geometries (channel cross-sectional area) and flow rate guided by the Hagen–Poiseuille equation
[31], with a lower shear rate generated through channels with a larger cross-sectional area. In cartilage and chondrocytes, abnormal fluid shear stress recapitulates the earmarks of OA, as indicated by the dysregulation of proinflammatory cytokines, matrix metalloproteinases, and pro-apoptotic factors
[33][34][35]. As such, the manipulation of shear stress for cartilage and bone OoC to mimic interstitial fluid flow was noted as early as 2006 for OA modelling. In the articular joint, a low fluid shear rate at 2–10 dynes cm
−2 [36], corresponding to interstitial flow, contributed to the chondroprotective effect on the cartilage tissue and promoted the ERK pathway, essential for osteoblast survival and proliferation
[34]. The perfusion of fluids exposes cells to shear stress that could be beneficial for chondrocyte regeneration
[37], inflammation response
[35], ECM synthesis
[35] as well as bone cell morphogenesis
[38]. Bao et al. subjected MSC-derived cartilage cells cultured in microfluidic devices with shear stress akin to interstitial fluid flow within the cartilage space (0.05 dynes/cm
2)
[15]. It was noted that cartilage cultures exposed to low shear stress showed improved collagen II and aggrecan synthesis compared to static and 2D cultures.
Apart from liquid shear stress, cartilage and bone tissues are also subjected to consistent mechanical stress during joint motion. Various attempts have been reported to generate mechanical stress within microfluidic devices suitable for cartilage tissue models. For example, Paggi et al. used a pneumatic channel network alongside the cartilage chambers
[14]. The chondrocyte can be subjected to physiological periodical mechanical compression of the channel walls against the cartilage cultures mimicking mechanical stress during joint motion by controlling the air pressure in and out of the pneumatic network. The application of rhythmic compression stress is potentially advantageous as a model for aging and metabolic-centric OA pathophysiology. However, it is worth noting that strategies implementing mechanical stress remain reliant on the flexibility of PDMS, which limits the mode of device fabrication. Although the use of plastic and additive manufacturing is gaining momentum as an alternative for microfluidic devices, the development of biocompatible flexible plastics remains slow. One possible way to circumvent this issue is to rely on multi-material device assembly strategies (akin to scaffold devices) to integrate flexible PDMS.
4. Mimicking Biochemical Environment/Chemical Gradient
One of the hallmarks of OA disease pathology is the inflammation response to the cartilage and bone tissue in both acute and post-traumatic injury. During acute joint damage, the regulation of cytokines, such as IL-1, IL-6, and IL-10, within the joint synovium affects the regulation of cartilage metabolism
[39]. In the context of cartilage, tuning both the growth factors and cytokines is crucial to the differentiation process, wound healing
[40], and the inflammation response of post-traumatic OA cases. Apart from cytokines, extracellular vesicles (EVs) within the synovial joint are known to regulate cartilage and joint tissue functions
[41]. Emerging evidence has indicated that synovium and or articular cartilage-secreted EVs carrying a wide variety of biomolecules, including mRNA, protein, microRNA, and inflammatory cytokines, are found to facilitate tissue cross-talk during OA
[42]. For instance, Wu et al. demonstrated that the subchondral osteoblast from sclerotic regions of OA patients secreted EVs containing miR-210-5p that is responsible for manipulating chondrocyte phenotype by altering the expression of ECM production and catabolic activities in vitro, and this miR expression was significantly enhanced in OA patients
[43]. Another study revealed that upregulation of long non-coding RNA plasmacytoma variant translocation 1 (PVT1) in isolated EVs from whole blood of OA patients induced apoptosis, inflammation responses and collagen degradation of chondrocytes by modulating the Toll-like receptor 4/NF-κB pathway through an miR-93-5p/HMGB1 axis
[44]. It is, therefore, highly desirable to develop a physiologically similar culture model that can potentially be used to examine the transport and uptake of the secreted EVs. The growing interest in utilising EVs for OA therapy would require a culture platform that can mimic a patient’s native biochemical environment as well.
Liquid mixing within microfluidic devices remains diffusion dominant due to the inherently low Re number (<100). It is common for microfluidic channels to contain flow dividers separating fluid streams to generate a chemical gradient, subjecting cells to a range of chemical concentrations depending on their position. Several microfluidic devices use this mechanism to generate polarised tissue cultures
[11] and perform dose-response studies for drugs
[32]. These devices typically consisted of two separate inlets (one for the drug/chemical of choice and the other as diluent), linked by branches downstream of the inlet, generating a range of chemical concentrations
[45]. Uniform mixing at the end of each flow outlet can be achieved by creating micro architectures or changing channel geometries to induce streamlined mixing
[32]. A controlled, non-uniform distribution of cytokines and growth could help for zonal differentiation of both chondrocyte and osteocyte phenotype—vital for capturing different aspects of a joint
[46] within one device.
To date, several demonstrations have integrated these strategies to tune growth factor exposure to the cartilage culture for OA models, which was covered extensively in the previous review by Banh et al.
[3]. For example, Tian et al. incorporated a concentration generator upstream of chondrocyte cultures to investigate interleukin-mediated chondrocyte proliferation parallelly. Within one setup, Tian et al. demonstrated the capability to screen multiple dosages of interleukin to induce cartilage repair
[45]. However, the footprint of these microfluidic devices will be relatively large due to the need to incorporate branched fluid channels and mixing mechanisms. In addition to a larger OoC footprint, for finer control of concentration, finer placement of flow dividers at 10 µm resolution might be needed
[45], which limits the device design to microfabrication. An alternative is to leverage hydrogels as a diffusion barrier to control chemokine distribution on-chip
[47], achievable by loading hydrogels into a microfluidic channel to form a diffusive barrier. The patterning of the hydrogel can be achieved by manipulating microchannel geometries or micropillar arrays
[48] (discussed in an earlier section). The hydrogel diffusion barrier porosity can be tuned by controlling the concentration of the hydrogel
[49] and the crosslinking process for controlling diffusion rates of essential biomolecules
[50]. This strategy has been highly successful in partitioning culture media content
[51], EVs
[52], and oxygen content as well
[8].
5. Localised Control of Oxygen Tension
In addition to controlling soluble growth factors, drugs, and cytokines, there is a need to tune the dissolved oxygen environment for OA. The mimicry of the hypoxia region in microfluidic joint models is particularly important since healthy articular chondrocytes rely on hypoxia to retain their metabolism and matrix synthesis
[53][54]. Evidence in human cartilage samples showed HIF-1 and HIF-2 regulate chondrogenesis and cartilage catabolism through the metalloproteinases and VEGF pathway
[55]. Hypoxia also helps regulate cartilage matrix catabolic enzymes in bone marrow-derived mesenchymal stem cells
[56]. The ability to model oxygen tension on-chip will be pivotal in enabling the setup of OA disease models and as a screening platform for potential stem cell therapies. Few approaches to date have been implemented to control the oxygen tension for chondrocyte culture on-chip, as most hypoxia environments were established under hypoxic incubators
[12][24]. While this approach effectively mimics the physiological oxygen tension in vivo, it remains challenging to model the tissue cross-talk of OA pathophysiology where co-culture with mildly hypoxic bone tissues is needed
[57]. As a result, there is a need for joint OoC to segregate the microenvironment for both cartilage and bone tissue while allowing small molecule diffusion between these tissue culture chambers.
Strategies for generating hypoxia locally for OoC tissue cultures remain limited
[54], with the earliest OA-related OoC work reported in 2021
[8]; this is partially due to the high technical barriers to implementing these platforms into tissue culture works, as many rely on highly specialised microfabrication techniques. However, techniques demonstrated with gut-on-chip, which utilise low oxygen media (generated by flowing media through a low oxygen chamber) or chemical scavengers, can be implemented for OA tissue co-culture with low technical entry barriers. For example, Ingber’s team demonstrated a gut-on-chip device with two separate media inlets (one with low oxygen medium using nitrogen and one with normoxic medium), which feeds into two closely separated culture compartments, creating a culture environment with heterogeneous oxygen tension mimicking the gut physiology
[57]. The chemical scavenging approach of controlling oxygen tension was also noted when Kim et al. utilised the perfusion of chemical oxygen scavengers to purge the oxygen level for gut-on-chip models
[58].
6. Mimicking Bone Cartilage Cross-Talk
Various OoC co-culture models were established to capture the cross-talk between chondrocytes, osteoblasts, osteoclasts, and macrophages as a potential OA model (
Table 1). With careful channel design geometry, it is possible to create a simplistic co-culture chip that can model tissue cross-talk with their physiological tissue structure
[59][60][61]. For example, Oliveria et al. demonstrated a co-culture platform for chondrocytes and monocytes to model the inflammation response and recruitment of monocyte during OA progression
[19]. Similar work was shown as well with bone–monocyte cultures
[62], synoviocyte–bone
[22], and synoviocyte–cartilage
[31] co-culture models. In general, approaches in microfluidic co-cultures for OA models encompass partitioned microchambers to segregate the individual cell types with a shared media circulating the chambers for cell–cell cross-talk. Multi-chamber OoCs offer a unique solution for pairing different cell cultures, useful to identify the mechanisms of complex diseases such as OA. For example, cross-talk between bone and cartilage, modelling for cartilage response to inflamed bone has been captured in multiple bone cartilage OoC models
[51]. Apart from bone and cartilage cross-talk contributing to OA disease progression, pathological changes in synovium have been associated with pain and disease progression in patients
[63][64].
Table 1. Reported OoC co-culture catering for the modelling of OA.
Synoviocytes have been found to play a key role in regulating OA inflammation cascade via proinflammatory factors such as nitric oxide and prostaglandin E
[67]. Inflammatory molecules, including IL-1β, TNF-α, and IL-6, expressed by activated macrophage-like synoviocytes, contribute to cartilage damage and bone alterations
[68]. At the same time, fibroblast-like synoviocytes from the site of joint pain in knee OA patients exhibit a distinct subset that plays a key role in promoting fibrosis, inflammation, and the growth and activity of neurons
[69]. These findings highlighted possible cross-talk between synovium and other tissues in the joint during OA.
Therefore, the pairing of synoviocytes and chondrocytes in OoCs would enable mechanistic investigations into inflammation regulation within the synovium. For example, Rothbauer et al. co-cultured synoviocyte and chondrocyte connected by a perfusion channel
[14] to study the paracrine signalling effect of synoviocyte on the sensitivity of chondrocyte to proinflammatory molecules.
OoC co-cultures of bone/cartilage with mesenchymal stem cells (MSCs) can be a potential application to identify stem cell-based therapies. MSC’s immunomodulatory role through EVs has been suggested for application as a treatment option for OA
[70][71]. At the time of writing, limited multi-cell co-culture with stem cells on-chips for OA had been observed
[55]. Since most MSCs and stem cell cultures are characterised by conventional 2D cultures, it is likely that differentiation and culture of stem cells with controlled microenvironments need to be thoroughly characterised before their co-culturing with other cell types on-chip.
Neovascularisation during OA involves multiple cell types (chondrocyte, osteoblast, and endothelial cells), which can potentially be studied in OoC co-cultures. The neovascularisation process is known to be assisted through VEGF signalling by the osteoblast
[72][73]. In addition to osteoblasts, endothelial cells play a significant role in OA neovascularisation through the digestion of the cartilage and inducing vascular formation through the VEGF signalling pathway
[72]. The development of microfluidic neovascularisation models largely focuses on other tissue types, as covered in existing reviews
[74]. Generally, for neovascularisation, co-culture of endothelium and fibroblasts are required. For example, Jeon’s team patterned HUVECS and fibroblasts in concentric cylinders creating vascular networks
[75]. Due to OA neovascularisation involving multiple cell types, modelling in OA angiogenesis would require co-culture with more than three cell types.
There is minimal control of the biochemical environment in many of these models due to the circulation of shared culture media within the culture device (Table 1).