Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hui-Ching Wang | -- | 3834 | 2023-03-07 02:49:36 | | | |
| 2 | Camila Xu | Meta information modification | 3834 | 2023-03-07 03:02:39 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Yeh, T.; Luo, C.; Du, J.; Huang, C.; Wang, M.; Chuang, T.; Gau, Y.; Cho, S.; Liu, Y.; Hsiao, H.; et al. Telomerase Reverse Transcriptase in Head and Neck Cancer. Encyclopedia. Available online: https://encyclopedia.pub/entry/41915 (accessed on 07 February 2026).
Yeh T, Luo C, Du J, Huang C, Wang M, Chuang T, et al. Telomerase Reverse Transcriptase in Head and Neck Cancer. Encyclopedia. Available at: https://encyclopedia.pub/entry/41915. Accessed February 07, 2026.
Yeh, Tsung-Jang, Chi-Wen Luo, Jeng-Shiun Du, Chien-Tzu Huang, Min-Hung Wang, Tzer-Ming Chuang, Yuh-Ching Gau, Shih-Feng Cho, Yi-Chang Liu, Hui-Hua Hsiao, et al. "Telomerase Reverse Transcriptase in Head and Neck Cancer" Encyclopedia, https://encyclopedia.pub/entry/41915 (accessed February 07, 2026).
Yeh, T., Luo, C., Du, J., Huang, C., Wang, M., Chuang, T., Gau, Y., Cho, S., Liu, Y., Hsiao, H., Chen, L., Pan, M., Wang, H., & Moi, S. (2023, March 07). Telomerase Reverse Transcriptase in Head and Neck Cancer. In Encyclopedia. https://encyclopedia.pub/entry/41915
Yeh, Tsung-Jang, et al. "Telomerase Reverse Transcriptase in Head and Neck Cancer." Encyclopedia. Web. 07 March, 2023.
Copy Citation
Telomerase, a specialized reverse transcriptase, is a large multi-subunit ribonucleoprotein complex that synthesizes telomeric DNA sequences and provides a molecular basis for unlimited proliferative potential. Several mechanisms could activate telomerase reverse transcriptase (TERT), the most common of which is promoter alternation. Two major hotspot TERT promoter mutations (C228T and C250T) have been reported in different malignancies such as melanoma, genitourinary cancers, CNS tumors, hepatocellular carcinoma, thyroid cancers, sarcomas, and HNCs. The frequencies of TERT promoter mutations vary widely across tumors and is quite high in HNCs (11.9–64.7%).
head and neck cancer
telomerase reverse transcriptase (TERT)
promoter mutations
1. Introduction
Head and neck cancers (HNCs) arising in the oral cavity, oropharynx, larynx, and hypopharynx were the seventh most common cancer worldwide in 2018 [1] and approximately seven to eight hundred thousand new cases are diagnosed each year worldwide [2]. The most common type, squamous cell carcinoma, is a highly lethal group of heterogeneous neoplasms often diagnosed at an advanced stage [3]. Tobacco and alcohol consumption are the main etiological factors [4]. Betel quid chewing and infection by oncogenic human papillomavirus (HPV) types 16 and 18 have emerged as important etiological factors for a subset of HNCs in the oral cavity and oropharynx, respectively [5][6][7][8][9][10]. HPV-positive malignancies represent 5–20% of all HNCs and 40–90% of those arising from the oropharynx [11]. The prevalence of HPV-driven HNCs has been dramatically increasing in developed countries, predominantly affecting middle-aged white men, non-smokers, non-drinkers, or mild-to-moderate drinkers with a higher socioeconomic status and better performance statuses than those with HPV-unrelated SCCs [11][12]. The treatment of HNC is generally multimodal, including surgery, chemotherapy, and radiotherapy, and differs according to disease stage, anatomical location, and surgical accessibility. However, despite significant progress in all therapeutic modalities, the 5-year overall survival (OS) rate of HNC patients remains unsatisfactory [13][14][15].
In the era of biomarker-driven personalized cancer therapy, several biomarkers have been proposed as prognostic and predictive factors in different cancers, such as KIT mutations in gastrointestinal stromal tumors, EGFR mutations in lung cancer, and HER2 overexpression in breast cancer [16]. However, unlike other cancer types, there are limited predictive biomarkers and targeted therapies for HNCs [4][16][17][18]. With the development of advanced technical approaches, genome, and exome analyses have provided a comprehensive view of genetic alterations in HNC and uncovered potential new therapeutic opportunities [17][19][20][21][22][23][24]. In addition to commonly mutated genes, such as TP53, CDKN2A, CCND1, PIK3CA, and NOTCH1, telomerase reverse transcriptase (TERT) promoter mutations have been detected in a significant proportion of HNC patients [3][4][12][13][15][25][26][27][28][29]. TERT is located on chromosome 5p15.33 in humans and is an integral and essential part of the telomerase holoenzyme, which plays a key role in cancer formation. Mostly, telomerase activity was increased by upregulation of TERT expression via several genetic and epigenetic alterations, and TERT promoter mutations are known as the most important [30]. However, the incidence of TERT promoter mutations varies in the head and neck subsites, and the association between TERT promoter mutations and outcomes is unclear.
2. Telomeres and Telomerase in Normal Cells
Telomeres are the physical ends of eukaryotic linear chromosomes [31][32]. In human cells, telomeres are composed of TTAGGG nucleotide repeats with a 3′ single-stranded overhang, and the variation ranges from 3 to 20 kilobase pairs [12][33]. They are bound by a six-member protein complex known as shelterin. Telomeres cover the coding DNA at the end to avoid loss of genetic information in linear DNA, act as a cap to prevent degradation by a nucleolytic attack, and prevent aberrant activation of a DNA damage response (DDR), which could lead to inappropriate processing of telomeres and as sites for double-strand break repair [31][34][35].
Telomerase, a specialized reverse transcriptase, is a large multi-subunit ribonucleoprotein complex that synthesizes telomeric DNA sequences and provides a molecular basis for unlimited proliferative potential [36]. Telomerase comprises two major components: the telomeric RNA component (also known as the telomerase RNA component: TERC or TR) and the telomerase reverse transcriptase (TERT), which is encoded by the TERT gene. The TERT is located in the human chromosome band 5p15.33, and the TERC is located at 3q26.3 [37]. The TERC serves as the template for telomere hexamer repeat additions onto the DNA, and the TERT is responsible for the reverse transcribing hexamer repeats onto the chromosomal ends [33][38][39][40]. TERT expression is silenced during development, unlike the TERC and other constitutively expressed telomerase components [35].
Telomerase is a key telomere length maintenance mechanism and is present in germline, hematopoietic, stem, and other rapidly renewing cells [41]. However, in most normal somatic cells, telomerase activity is extremely low or absent. Therefore, loss of telomeric repeats occurs at each round of DNA replication, after which the telomeres are reduced to a critical length [42]. Critical telomere attrition elicits a DDR that mediates cell cycle arrest and leads to replicative senescence or apoptosis via the p53 or Rb tumor suppressor pathways [35][43]. Telomere attrition acts as a barrier to replicative immortality, also called a “mitotic clock” that limits the cell cycle number and further triggers cellular senescence [33][35]. Although rare, in the absence of telomerase, some cells employ another DNA recombination mechanism, termed alternative lengthening of telomeres (ALT), which reverses telomere attrition to bypass senescence [44].
3. Telomere and Telomerase in Cancer Cells
Telomere length and telomerase activity are crucial for cellular immortalization, tumorigenesis, and cancer progression. Telomere maintenance via telomerase reactivation is a nearly universal hallmark of cancer cells [35][45][46]. The vast majority of cancers overcome replicative senescence by upregulating TERT expression and telomerase activity [35]. Telomerase activity is upregulated in 80–90% of malignancies, enabling unlimited replication of cancer cells, similar to embryonic and stem cells [13][15][47]. For the remaining 10–15% of cancers, upregulation of telomerase activity is achieved through the ALT pathway [48].
There are several ways to upregulate telomerase activity and activate the normally silent human TERT (hTERT) gene. The mechanisms of hTERT activation include chromosomal rearrangements (i.e., duplications, amplifications, insertions, interchromosomal changes, inverted orientations, or deletions), TERT promoter somatic mutations, epigenetic modifications (i.e., DNA methylation, or post-transcriptional regulation by microRNAs), transcriptional activators or repressors, TERT gene polymorphism and alternative splicing (i.e., pre-mRNA alternative splicing of the TERT gene) [30][49][50][51]. In a pan-cancer genomics study, Barthel et al. detected TERT expression in 73% of the 6835 total tumor samples, which were associated with TERT point mutations, rearrangements, DNA amplifications, and transcript fusions. Among the TERT-expressing samples, there were 31% TERT promoter mutations, 3% TERT amplifications, 3% TERT structural variants, 5% TERT promoter structural variants, and 53% TERT promoter methylation [51]. Some of these mechanisms may interact with each other and have a synergistic effect on TERT expression [30].
Besides the canonical role of telomerase in telomere maintenance, there are also some non-canonical functions (telomere length-independent mechanisms) in tumorigenesis, such as the regulation of metabolic mechanisms, epigenetic regulation, and modulation of chromatin, oxidative stress protection, RNA silencing, signal transduction pathways (Wnt and c-MYC signaling pathways), enhanced mitochondrial function, cell adhesion, and migration [30][52][53][54][55][56][57][58].
4. Telomerase Reverse Transcriptase (TERT) Promoter Mutations
Among the several mechanisms of hTERT activation, TERT somatic promoter mutations are the most common non-coding driver mutations in cancer [30][59] and occurred at a high frequency in over 50 cancer types [60]. They have been reported in two major hotspots (mainly C ˃ T transitions), which are located at −124 and −146 base pairs upstream of the transcriptional start site on chromosome 5 and are designated as C228T and C250T, respectively [13][25][32][61][62]. A less frequent hTERT promoter mutation −57 base pairs upstream of the transcriptional start site with an A > C transition (at position 1,295,161 on chromosome 5) has been found to be a disease-segregating germline mutation in a melanoma-prone family [63]. Other less frequent, yet recurrent, mutations on chromosome 5 have also been discovered in cancers at the following positions: 1,295,228 C > A, 1,295,248–1,295,243 CC > TT, and 1,295,161 A > C [64].
TERT somatic promoter mutations are predominantly heterozygous and lead to the generation of an 11 bp sequence, CCCGGAAGGGG, which is similar to the E26 transformation-specific (ETS) factor binding motif [60][65]. Then, ETS binding factors, such as GA-binding protein (GABP), are recruited. This recruitment resulted in direct transcriptional activation of hTERT expression and promoted an epigenetic shift from a repressed to active chromatin conformation [35][65][66][67][68]. These promoter mutations were proven to be associated with higher levels of TERT mRNA, TERT protein, telomerase enzymatic activity, and telomere length in a study of 23 human urothelial cancer cell lines [69].
Two TERT promoter hotspot mutations, C228T and C250T are the most common; however, their frequencies vary widely across tumors from different sites (Table 1). These mutations occur most frequently in cancers with low rates of self-renewal [25] and are rare in pediatric and young adult cancers [3][70]. The highest frequencies of TERT promoter mutations have been reported in melanoma, bladder cancer, urothelial carcinoma, CNS tumors, hepatocellular carcinoma, thyroid cancer, basal cell carcinoma, and cutaneous squamous cell carcinoma. Due to the variety of sarcoma subtypes, the prevalence of TERT promoter mutations varies widely, and the highest TERT promoter mutation rate is reported in myxoid liposarcoma (79.1%) [25].
Table 1. Frequency spectrum of hTERT promoter mutations across different cancer types.
| Cancer Type | Mutation Frequency (%) | Reference |
|---|---|---|
| Malignant melanoma | 17.0–85.0 | [61][63][71][72] |
| Genitourinary cancers | ||
| Bladder cancer | 59.0–85.0 | [25][61][73][74][75][76] |
| Urothelial carcinomas | 29.5–64.5 | [77][78] |
| Kidney cancers | 0 | [61] |
| Prostate Cancer | 0 | [79] |
| CNS tumors | ||
| Glioblastoma | 54.0–84.0 | [61][70][73][78][80] |
| Other gliomas (ependymoma, astrocytoma, mixed glioma, oligodendroglioma) | 2.7–78.0 | [25][64][70][78] |
| Medulloblastoma | 33.3–65.0 | [70][78] |
| Hepatocellular carcinoma | 31.4–59.0 | [25][78][81][82][83][84] |
| Thyroid cancer (papillary, follicular, poorly differentiated, and anaplastic carcinomas) | 3.4–46.3 | [61][85][86][87] |
| Gastrointestinal stromal tumor | 0–3.8 | [61][88] |
| Malignant pleural mesothelioma | 11.3 | [89] |
| Atypical fibroxanthomas | 93.0 | [90] |
| Sarcomas (chondrosarcoma, fibrosarcoma, myxofibrosarcoma, myxoid liposarcoma, osteosarcoma, pleomorphic dermal sarcomas) | 4.3–79.1 | [25][90][91] |
| Basal cell carcinoma of the skin | 73.8 | [92] |
| Squamous cell carcinoma of the skin | 20.0–74.0 | [25][92][93] |
| Squamous cell carcinoma of esophageal | 1.6 | [94] |
| Squamous cell carcinoma of penile | 48.6 | [95] |
| Squamous cell carcinoma of the head and neck | 11.9–64.7 | [3][4][13][15][25][26][27][28][29][32][93][96][97] |
| Squamous cell carcinoma of the cervix | 0–21.4 | [25][26][93][96] |
| Breast cancer, colorectal cancer, ovarian cancer, esophageal adenocarcinoma, acute myeloid leukemia, chronic lymphoid leukemia, pancreatic cancer, and testicular carcinoma | 0–5.0 | [61][78] |
5. TERT Promoter Mutations in Head and Neck Squamous Cell Carcinoma
5.1. The Frequency of TERT Promoter Mutations
For HNCs, the frequency of TERT promoter mutations varied significantly among previous studies. These differences could be explained by the tumor subsite, sample size, methodological sensitivity, risk factors, and population ethnicity (Table 2).
Table 2. Summary of studies evaluating the association between head and neck cancers with TERT promoter mutations.
| Author, Country (Year) | Case Numbers | Cancer Sites | Prevalence of TERT Promoter Mutations | Special Findings | The Association with Survival |
|---|---|---|---|---|---|
| Killela, USA (2013) [25] |
70 | 31 Oral cavity 23 Oropharynx 4 Supraglottic 12 Others |
Total: 17.1% (12/70) C228T: 14.8% C250T: 2.8% |
Highest frequency in tongues (47.8%, 11/23) | N/A |
| Schwaederle, USA (2018) [32] |
28 | 28 HNC | Total: 28.6% (8/28) | N/A | Trend toward shorter survival |
| Cheng, USA (2015) [93] |
12 | 12 HNSCC | Total: 16.67% (2/12) C228T: 16.67% C250T: 0% |
No significant correlation was observed. | N/A |
| Barczak, USA (2017) [15] |
61 | 25 Mouth 25 Voice box 5 Nose/sinuses 6 Throat |
C250T homozygous T/T allele: 36% heterozygous C/T allele: 26% |
Homozygous T/T mutation is associated with the grade of the tumor. | N/A |
| Yu, USA (2021) [29] |
117 | 74 Oral cavity 24 Larynx 5 Hypopharynx 14 HPV (-) oropharynx |
Total: 53.8% (63/117) C228T: 33.3% C250T: 9.4% C250T, C254T: 6% C228A: 4.3% CC434TT: 0.9% |
Highest frequency in the oral cavity (81.1%, 60/74) | Increased risk of locoregional failure, but not distant failure or OS. |
| Morris, USA (2017) [97] |
53 | 20 Oral cavity 18 Oropharynx 7 Larynx 2 Hypopharynx 6 Others (4 sinonasal cavity) |
Total: 32.1% (17/53) C228T: 20.8% C250T: 5.7% C228A: 1.9% |
TERT mutation and HPV infection may represent parallel mechanisms. | N/A |
| Boscolo-Rizzo, Italy (2020) [3] |
101 | 27 Oral cavity 23 Oropharynx 15 Hypopharynx 36 Larynx |
Total: 11.9% (12/101) C228T: 9.9% C250T: 2% |
Highest frequency in the oral cavity (37%) TERT levels did not significantly differ according to the mutational status of TERT promoter. |
No significant association between TERT promoter status and OS. Higher TERT levels, worse OS (43.6% vs. 60.1%) |
| Annunziata, Italy (2018) [96] |
24 | 15 Oral cavity 9 Oropharynx |
Total: 37.5% (9/24) C228T: 8.3% C250T: 12.5% Other: 16.7% |
No mutation in oropharynx cancer. Mutations were independent of HPV status. |
N/A |
| Yilmaz, Turkey (2020) [4] |
189 | 102 Oral cavity 22 Oropharynx 6 Hypopharynx 59 Larynx |
Total: 43.9% (83/189) C228T: 29.6% C250T: 11.6% C228A: 2.6% |
Highest frequency in the oral cavity (75.5%, 77/102). TERT mutations are associated with younger age, female gender, and an inverse relationship to smoking and alcohol consumption. |
No difference |
| Arantes, Brazil (2020) [13] |
88 | 69 Oral cavity 11 Larynx 8 Pharynx |
Total: 27.3% (24/88) C228T: 6.8% C250T: 20.5% |
94.4% C250T were alcohol consumers. 66.7% C228T were not alcohol consumers |
Decreased 5-year DFS and OS in C228T |
| Vinothkumar, India (2016) [26] |
41 | 41 Oral cavity | Total: 31.7% (13/41) C228T: 21.9% C250T: 9.7% |
No significant correlation was observed. | N/A |
| Chang, Taiwan (2017) [28] |
201 | 201 Oral cavity | Total: 64.7% (130/201) C228T: 51.7% C250T: 12.9% |
C228T mutation was associated with betel nut chewing. | No difference |
| Qu, China (2014) [27] |
235 | 235 Laryngeal | Total: 27% (64/235) C250T: 23.8% C228T: 3.4% |
Not significantly correlate with any clinicopathological variables | Poor survival, especially C250T mutation |
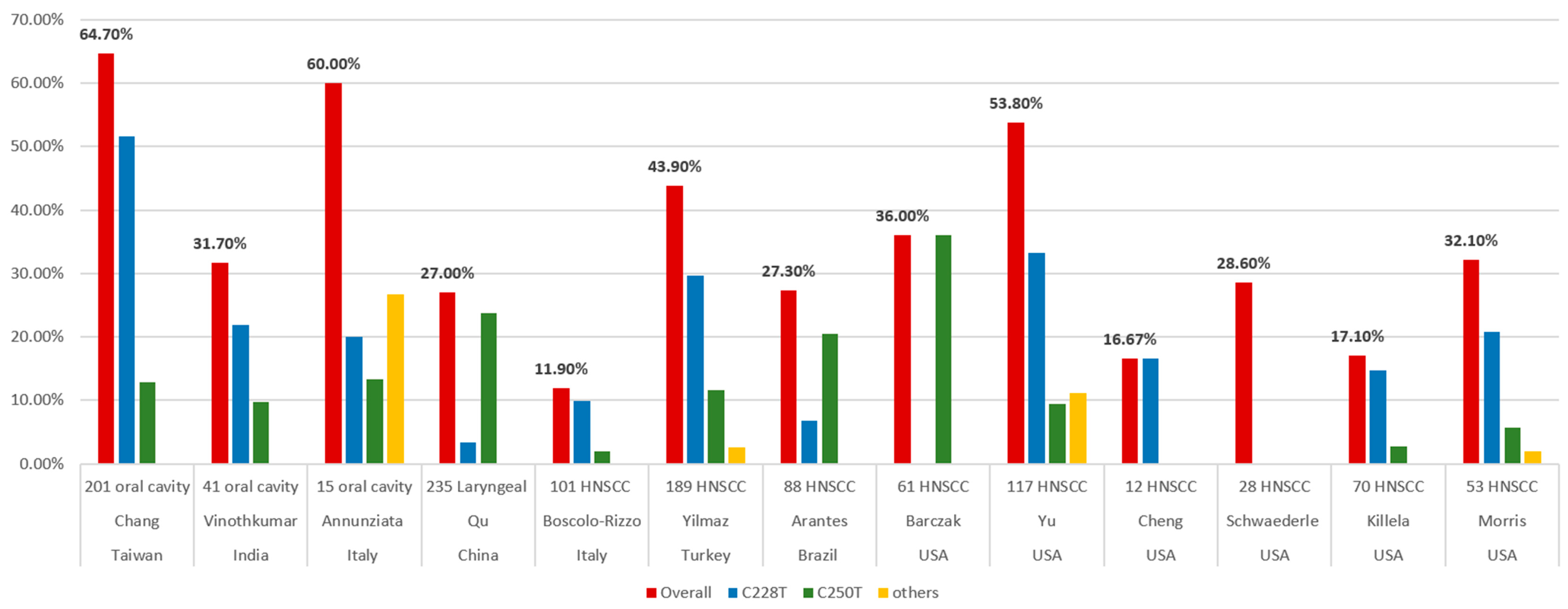
Killela et al. surveyed 70 oral cavity cancers and identified TERT promoter mutations in 12 of the tumors (17.1%) [25]. Schwaederle et al. analyzed 423 cases of TERT promoter alterations using next-generation sequencing (NGS). Only 28 patients (6.6%) had HNCs. The incidence of TERT promoter alternations was 14.4% (61 of 423) in the overall population and 28.6% (8 of 28) in HNCs [32]. Cheng et al. collected 84 cases of SCC from different sites, including 12 HNC and C228T mutations, which were detected in 16.67% (2 of 12) [93]. Barczak et al. analyzed 61 HNC patients to determine the prevalence of the hTERT promoter C250T mutation. High-resolution melting mutation analysis was used to identify the C250T hTERT promoter mutation, followed by sequencing verification in 10% of the samples. The prevalence of the hTERT promoter C250T mutation was 36% [15]. Yu et al. identified TERT promoter mutations in 117 patients with SCC of the oral cavity (N = 74), larynx (N = 24), hypopharynx (N = 5), and HPV-negative SCC of the oropharynx (N = 14) using NGS. Overall, 63 patients (53.8%) had TERT promoter alterations, and the most common mutations were C228T and C250T [29]. Morris et al. collected 53 patients and 20 oral cavities, 18 oropharynges, 7 larynges, and 2 hypopharynges. Overall, the frequency of the TERT alteration was 32.1% (17 of 53), yet it was much higher in HPV-negative tumors (53% vs. 4.3%). Remarkably, 91% (10 of 11) of the HPV-negative tongue SCCs possessed TERT mutations [97].
In Italy, Boscolo-Rizzo et al. analyzed cancer tissue and adjacent mucosa specimens from 101 patients with HNCs and evaluated the prevalence of the TERT promoter mutations by Sanger sequencing. The tumor subsites in the HNCs included the oral cavity (N = 27), oropharynx (N = 23), hypopharynx (N = 15), and larynx (N = 36). The TERT promoter harbored mutations in 12 tumors (11.9%), with C228T and C250T, which accounted for 83.3% and 16.7%, respectively. They also evaluated the TERT mRNA level and found no significant difference between the TERT mRNA level and the mutational status of the TERT promoter [3]. Annunziata et al. analyzed tumor biopsies from 15 oral SCCs and nine oropharyngeal SCCs. The frequency of TERT promoter mutations was 60% (9 of 15) in oral SCCs and was absent in oropharyngeal SCCs. There were five hotspot mutations (three C228T and two C250T) and four other mutations. They also investigated the TERT mRNA levels and identified that the TERT mRNA levels were comparable to those detected in peri-tumor tissues. However, these data were from six oropharyngeal SCCs and illustrated that they all lacked mutations in the TERT promoter [96].
In Turkey, Yilmaz et al. collected a total of 189 patients with HNCs, including 102 oral cavities, 22 oropharynges, 6 hypopharynges, and 59 larynges. The TERT gene expression was examined by polymerase chain reaction (PCR)-based direct sequencing. TERT promoter mutations were detected in 43.9% (83 of 189) of the cases. Three TERT promoter region mutations were detected: C228T (56 of 83; 67.5%), C250T (22 of 83; 26.5%), and C228A (5 of 83; 6%). The frequency of the C228T mutation was almost twice that of the C250T and C228A mutations [4]. In Brazil, Arantes et al. collected 88 HNC patients and analyzed the TERT promoter mutations C228T and C250T using pyrosequencing. The overall prevalence of the TERT hotspot mutations is 27.3% (6.8% at locus C228T and 20.5% at C250T) [13]. In India, Vinothkumar et al. analyzed 181 primary tumors of the uterine cervix and oral cavity using PCR amplification and sequencing. A high frequency of TERT hotspot mutations was observed in both cervical (30 of 140, 21.4%) and oral (13 of 41, 31.7%) SCCs. Among the oral cancer samples, the TERT promoter hotspot mutations were frequent, while the C228T mutation (69.2%) was twice as frequent as the C250T (30.8%) [26].
In Taiwan, Chang et al. included 201 oral cavity SCC tumors and adjacent normal tissues to detect two TERT promoter mutations (C228T and C250T) using Sanger sequencing. Overall, the TERT hotspot promoter mutations occurred at a high frequency (64.7%) in patients with oral cavity SCCs. There were 52.5% (104 of 201) and 12.9% (26 of 201) oral cavity SCC tumor tissues containing that contained the C228T and C250T mutations, respectively [28].
In China, Qu et al. obtained 235 laryngeal cancer tissues using a pyrosequencing assay to detect the TERT promoter mutations C228T and C250T. The TERT promoter hotspot mutations were present in 27% (64 of 235) of the samples. The TERT C250T mutations were more common (56 of 235) than the C228T mutations (8 of 235) [27]. In Figure 1, researchers summarized the reported frequencies of the TERT promoter mutations in HNCs from the various studies mentioned above.

Figure 1. Frequencies of TERT promoter mutations in head and neck cancers from different studies.
5.2. TERT Promoter Mutations in Different Anatomic Distribution
HNC is a heterogeneous group of tumors involving distinct anatomical sites and subsites with varying etiological factors. Yu et al. showed that TERT promoter mutations were more abundant in oral cavity SCCs than in laryngopharyngeal cancers (81.1% vs. 7.0%) [29]. Boscolo-Rizzo et al. demonstrated that the prevalence of TERT hotspot promoter mutations is significantly higher in oral cavity SCCs (37%) [3]. Annunziata et al. also showed that TERT promoter mutations were predominant in oral SCCs (60%), yet absent in oropharyngeal SCCs [96]. Arantes et al. reported that 92% of the mutation cases were located in the oral cavity [13]. Finally, Yilmaz et al. showed that the frequency of the TERT promoter mutations in oral SCCs (75.5%) was significantly higher than in the other locations [4]. The anatomic distribution of cases is strongly associated with TERT promoter mutations, and the highest frequency is in oral cavity cancers.
As for the subsites in oral cavity SCCs, Arantes et al. noticed that 92% of the mutated cases were mainly in the tongue [13]. Killela et al. also revealed that 11 out of the 12 cancers with TERT promoter mutations were in the oral tongue, although only 23 of the 70 oral cavity cancers originated in the oral tongue [25]. However, Yilmaz et al. demonstrated that the highest rate was related to the buccal location and the lowest to the floor of the mouth (82.35% and 61.53%, respectively), although the difference was not statistically significant [4].
5.3. TERT Promoter Mutation and Human Papillomavirus Status
An association between HPV infection and oropharyngeal SCC has been proven. It was also clear that the molecular landscape and clinical pattern were different between HPV-positive and HPV-negative oropharyngeal cancers [10]. Only two studies have investigated the association between HPV status and TERT promoter mutations.
In a cohort of 53 patients with advanced HNCs, performed by Morris et al., a very high TERT alternation rate (53%, 16 of 30) was present in 30 HPV-negative tumors, however, there was only one TERT alternation (4.3%), which was a TERT amplification rather than a hotspot mutation, in 23 HPV-positive tumors. HPV-negative tongue SCCs showed the highest TERT mutation rate (91%). This demonstrated that TERT mutations and HPV infection may represent parallel mechanisms of telomerase activation in HNCs [97]. In another cohort study conducted by Annunziata et al., among the 9 patients with TERT promoter mutations in 15 oral SCC patients, 7 were HPV-negative and 2 were HPV-positive (p = 0.486). The frequency of TERT mutations was also independent of HPV tumor status in oral cancer [96].
5.4. TERT Promoter Mutation and Tobacco, Alcohol, and Betel Quid
Aside from HPV infection, tobacco smoking, alcohol consumption, and betel quid chewing are the other three main etiological factors of HNC [4][5]. Until now, the relationship between the TERT promoter mutations and these three factors remains inconclusive.
In a Brazilian cohort of 88 patients with HNC conducted by Arantes et al., the frequency of the C250T mutation appeared to be higher in alcohol consumers. Of the patients harboring the TERT promoter mutation C250T, 94.4% were alcohol consumers, and 66.7% of the patients harboring the TERT promoter mutation C228T did not consume alcohol [13]. In a Chinese cohort of 235 laryngeal cancer cases reported by Qu et al., hotspot mutations were not significantly correlated with any clinicopathological variables. However, TERT promoter mutations, particularly the C250T mutation, were more frequent in smoking patients (47 of 130) than in non-smoking patients (9 of 49), although no statistical significance was noted [27]. In a cohort of 201 patients with oral cavity SCC performed by Chang et al. in Taiwan, the C228T mutation was significantly associated with betel nut chewing [28]. In contrast, in a Turkish cohort of 189 HNC patients performed by Yilmaz et al., TERT promoter region mutations in HNC were inversely related to smoking and alcohol consumption [4].
5.5. TERT Promoter Mutation and Other Factors
Schwaederle et al. demonstrated that TERT promoter alterations are more frequent in men. They were also associated with brain cancers, skin/melanoma, head, and neck tumors, and increased median numbers of alterations in the univariate analysis. However, this association in head and neck tumors was not found in further multivariate analyses [32]. Yilmaz et al. reported that TERT promoter region mutations in HNCs are associated with younger age and female genders in a cohort from Turkey [4]. Barczak et al. demonstrated a significant association between the frequency of the homozygous C250T mutation and tumor grade (T1 = 27%, T2 = 36%, T3 = 35%, T4 = 46%, p ≤ 0.0001) [15]. However, in a cohort of 41 patients with oral SCCs, performed by Vinothkumar et al. in India, no significant correlation was observed between any of the genotypes and the clinicopathological characteristics [26].
5.6. TERT Promoter Mutation and Survival
TERT promoter mutations in various reports of different cancers have been associated with aggressive characteristics, poor outcomes, and shorter survival [98][99][100][101]. In HNC, Qu et al. showed that TERT promoter mutations significantly affected the overall survival of laryngeal cancer patients, particularly those with the C250T mutation. TERT promoter mutations were significant predictors of poor prognosis in patients with laryngeal cancer, as an independent variable, with respect to age, tumor localization, TNM stage, tumor invasion, lymph node metastasis, and smoking history [27]. Schwaederle et al. also demonstrated a significantly shorter overall survival in patients harboring the TERT promoter alterations in the overall population in a univariate analysis. Subanalyses of the three tumor types with the highest prevalence of TERT alterations consistently showed a trend toward shorter survival for patients with altered TERT promoters in brain tumors, head, and neck cancers, and melanoma/skin tumors [32]. Arantes et al. demonstrated no statistically significant association between the presence of hotspot mutations (C228T and C250T) and survival. However, the presence of the C228T mutation impacted patient outcomes, with a significant decrease in 5-year disease-free survival (20.0 vs. 63.0%) and 5-year overall survival (16.7 vs. 45.1%) [13].
Similar results were reported by Yu et al. [29]. They reported that the TERT promoter mutations were associated with locoregional failure (LRF) in the overall cohort and in oral cavity SCCs. This increased risk for LRF is independent of the oral cavity primary site, TP53 mutation status, extracapsular extension, and positive surgical margins suggesting that the TERT promoter mutations are an independent biomarker of LRF rather than a surrogate for OSCCs, or other known prognostic markers. The cumulative incidence of LRF was similar between the two types of TERT promoter mutations (C250T and C228A/T groups), and both were associated with a higher cumulative incidence of LRF compared to wildtype tumors. Overall, they demonstrated that TERT promoter mutations were associated with an increased risk of LRF, although not with distant failure or overall survival [29].
In contrast, Yilmaz et al. did not find a significant association between the presence of TERT mutations and OS, despite patients with HNCs harboring TERT mutations exhibiting a slightly shorter median OS [4]. Boscolo-Rizzo et al. showed no significant association between the TERT promoter status and overall survival, although the TERT mRNA level had an impact on clinical outcomes [3]. Chang et al. also reported that there was no significant difference in overall survival, disease-specific survival, and disease-free survival between TERT promoter mutations and the wildtype [28].
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34.
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424.
- Boscolo-Rizzo, P.; Giunco, S.; Rampazzo, E.; Brutti, M.; Spinato, G.; Menegaldo, A.; Stellin, M.; Mantovani, M.; Bandolin, L.; Rossi, M.; et al. TERT promoter hotspot mutations and their relationship with TERT levels and telomere erosion in patients with head and neck squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2020, 146, 381–389.
- Yilmaz, I.; Erkul, B.E.; Ozturk Sari, S.; Issin, G.; Tural, E.; Terzi Kaya Terzi, N.; Karatay, H.; Celik, M.; Ulusan, M.; Bilgic, B. Promoter region mutations of the telomerase reverse transcriptase (TERT) gene in head and neck squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2020, 130, 63–70.
- Chen, Y.J.; Chang, J.T.; Liao, C.T.; Wang, H.M.; Yen, T.C.; Chiu, C.C.; Lu, Y.C.; Li, H.F.; Cheng, A.J. Head and neck cancer in the betel quid chewing area: Recent advances in molecular carcinogenesis. Cancer Sci. 2008, 99, 1507–1514.
- Gooi, Z.; Chan, J.Y.; Fakhry, C. The epidemiology of the human papillomavirus related to oropharyngeal head and neck cancer. Laryngoscope 2016, 126, 894–900.
- Mallen-St Clair, J.; Alani, M.; Wang, M.B.; Srivatsan, E.S. Human papillomavirus in oropharyngeal cancer: The changing face of a disease. Biochim. Biophys. Acta 2016, 1866, 141–150.
- Lee, Y.A.; Li, S.; Chen, Y.; Li, Q.; Chen, C.J.; Hsu, W.L.; Lou, P.J.; Zhu, C.; Pan, J.; Shen, H.; et al. Tobacco smoking, alcohol drinking, betel quid chewing, and the risk of head and neck cancer in an East Asian population. Head Neck 2019, 41, 92–102.
- Hashibe, M.; Brennan, P.; Benhamou, S.; Castellsague, X.; Chen, C.; Curado, M.P.; Dal Maso, L.; Daudt, A.W.; Fabianova, E.; Fernandez, L.; et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: Pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J. Natl. Cancer Inst. 2007, 99, 777–789.
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282.
- Boscolo-Rizzo, P.; Del Mistro, A.; Bussu, F.; Lupato, V.; Baboci, L.; Almadori, G.; MC, D.A.M.; Paludetti, G. New insights into human papillomavirus-associated head and neck squamous cell carcinoma. Acta Otorhinolaryngol. Ital. Organo Uff. Della Soc. Ital. Di Otorinolaringol. E Chir. Cervico-Facciale 2013, 33, 77–87.
- Boscolo-Rizzo, P.; Da Mosto, M.C.; Rampazzo, E.; Giunco, S.; Del Mistro, A.; Menegaldo, A.; Baboci, L.; Mantovani, M.; Tirelli, G.; De Rossi, A. Telomeres and telomerase in head and neck squamous cell carcinoma: From pathogenesis to clinical implications. Cancer Metastasis Rev. 2016, 35, 457–474.
- Arantes, L.; Cruvinel-Carloni, A.; de Carvalho, A.C.; Sorroche, B.P.; Carvalho, A.L.; Scapulatempo-Neto, C.; Reis, R.M. TERT Promoter Mutation C228T Increases Risk for Tumor Recurrence and Death in Head and Neck Cancer Patients. Front. Oncol. 2020, 10, 1275.
- Carvalho, A.L.; Nishimoto, I.N.; Califano, J.A.; Kowalski, L.P. Trends in incidence and prognosis for head and neck cancer in the United States: A site-specific analysis of the SEER database. Int. J. Cancer 2005, 114, 806–816.
- Barczak, W.; Suchorska, W.M.; Sobecka, A.; Bednarowicz, K.; Machczynski, P.; Golusinski, P.; Rubis, B.; Masternak, M.M.; Golusinski, W. hTERT C250T promoter mutation and telomere length as a molecular markers of cancer progression in patients with head and neck cancer. Mol. Med. Rep. 2017, 16, 441–446.
- Kang, H.; Kiess, A.; Chung, C.H. Emerging biomarkers in head and neck cancer in the era of genomics. Nat. Rev. Clin. Oncol. 2015, 12, 11–26.
- The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582.
- Juodzbalys, G.; Kasradze, D.; Cicciù, M.; Sudeikis, A.; Banys, L.; Galindo-Moreno, P.; Guobis, Z. Modern molecular biomarkers of head and neck cancer. Part I. Epigenetic diagnostics and prognostics: Systematic review. Cancer Biomark. Sect. A Dis. Markers 2016, 17, 487–502.
- Ausoni, S.; Boscolo-Rizzo, P.; Singh, B.; Da Mosto, M.C.; Spinato, G.; Tirelli, G.; Spinato, R.; Azzarello, G. Targeting cellular and molecular drivers of head and neck squamous cell carcinoma: Current options and emerging perspectives. Cancer Metastasis Rev. 2016, 35, 413–426.
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.X.; Zhang, J.; Wang, J.; et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011, 333, 1154–1157.
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160.
- Parfenov, M.; Pedamallu, C.S.; Gehlenborg, N.; Freeman, S.S.; Danilova, L.; Bristow, C.A.; Lee, S.; Hadjipanayis, A.G.; Ivanova, E.V.; Wilkerson, M.D.; et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 15544–15549.
- Seiwert, T.Y.; Zuo, Z.; Keck, M.K.; Khattri, A.; Pedamallu, C.S.; Stricker, T.; Brown, C.; Pugh, T.J.; Stojanov, P.; Cho, J.; et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin. Cancer Res. 2015, 21, 632–641.
- Lechner, M.; Frampton, G.M.; Fenton, T.; Feber, A.; Palmer, G.; Jay, A.; Pillay, N.; Forster, M.; Cronin, M.T.; Lipson, D.; et al. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV-tumors. Genome Med. 2013, 5, 49.
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A., Jr.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026.
- Vinothkumar, V.; Arunkumar, G.; Revathidevi, S.; Arun, K.; Manikandan, M.; Rao, A.K.; Rajkumar, K.S.; Ajay, C.; Rajaraman, R.; Ramani, R.; et al. TERT promoter hot spot mutations are frequent in Indian cervical and oral squamous cell carcinomas. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016, 37, 7907–7913.
- Qu, Y.; Dang, S.; Wu, K.; Shao, Y.; Yang, Q.; Ji, M.; Shi, B.; Hou, P. TERT promoter mutations predict worse survival in laryngeal cancer patients. Int. J. Cancer 2014, 135, 1008–1010.
- Chang, K.P.; Wang, C.I.; Pickering, C.R.; Huang, Y.; Tsai, C.N.; Tsang, N.M.; Kao, H.K.; Cheng, M.H.; Myers, J.N. Prevalence of promoter mutations in the TERT gene in oral cavity squamous cell carcinoma. Head Neck 2017, 39, 1131–1137.
- Yu, Y.; Fan, D.; Song, X.; Zakeri, K.; Chen, L.; Kang, J.; McBride, S.; Tsai, C.J.; Dunn, L.; Sherman, E.; et al. TERT Promoter Mutations Are Enriched in Oral Cavity Cancers and Associated With Locoregional Recurrence. JCO Precis. Oncol. 2021, 5, 1259–1269.
- Dratwa, M.; Wysoczańska, B.; Łacina, P.; Kubik, T.; Bogunia-Kubik, K. TERT-Regulation and Roles in Cancer Formation. Front. Immunol. 2020, 11, 589929.
- Giardini, M.A.; Segatto, M.; da Silva, M.S.; Nunes, V.S.; Cano, M.I. Telomere and telomerase biology. Prog. Mol. Biol. Transl. Sci. 2014, 125, 1–40.
- Schwaederle, M.; Krishnamurthy, N.; Daniels, G.A.; Piccioni, D.E.; Kesari, S.; Fanta, P.T.; Schwab, R.B.; Patel, S.P.; Parker, B.A.; Kurzrock, R. Telomerase reverse transcriptase promoter alterations across cancer types as detected by next-generation sequencing: A clinical and molecular analysis of 423 patients. Cancer 2018, 124, 1288–1296.
- Bhari, V.K.; Kumar, D.; Kumar, S.; Mishra, R. Shelterin complex gene: Prognosis and therapeutic vulnerability in cancer. Biochem. Biophys. Rep. 2021, 26, 100937.
- Rhodes, D.; Fairall, L.; Simonsson, T.; Court, R.; Chapman, L. Telomere architecture. EMBO Rep. 2002, 3, 1139–1145.
- Guterres, A.N.; Villanueva, J. Targeting telomerase for cancer therapy. Oncogene 2020, 39, 5811–5824.
- Cong, Y.S.; Wright, W.E.; Shay, J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. MMBR 2002, 66, 407–425.
- Cao, Y.; Bryan, T.M.; Reddel, R.R. Increased copy number of the TERT and TERC telomerase subunit genes in cancer cells. Cancer Sci. 2008, 99, 1092–1099.
- Lu, W.; Zhang, Y.; Liu, D.; Songyang, Z.; Wan, M. Telomeres-structure, function, and regulation. Exp. Cell Res. 2013, 319, 133–141.
- McKelvey, B.A.; Umbricht, C.B.; Zeiger, M.A. Telomerase Reverse Transcriptase (TERT) Regulation in Thyroid Cancer: A Review. Front. Endocrinol. 2020, 11, 485.
- Greider, C.W.; Blackburn, E.H. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 1987, 51, 887–898.
- Shammas, M.A. Telomeres, lifestyle, cancer, and aging. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 28–34.
- Yuan, X.; Larsson, C.; Xu, D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: Old actors and new players. Oncogene 2019, 38, 6172–6183.
- Jacobs, J.J.; de Lange, T. Significant role for p16INK4a in p53-independent telomere-directed senescence. Curr. Biol. CB 2004, 14, 2302–2308.
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016, 8, 69.
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674.
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46.
- Chen, C.H.; Chen, R.J. Prevalence of telomerase activity in human cancer. J. Formos. Med. Assoc. 2011, 110, 275–289.
- Cesare, A.J.; Reddel, R.R. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 2010, 11, 319–330.
- Shay, J.W. Are short telomeres predictive of advanced cancer? Cancer Discov. 2013, 3, 1096–1098.
- Leão, R.; Apolónio, J.D.; Lee, D.; Figueiredo, A.; Tabori, U.; Castelo-Branco, P. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: Clinical impacts in cancer. J. Biomed. Sci. 2018, 25, 22.
- Barthel, F.P.; Wei, W.; Tang, M.; Martinez-Ledesma, E.; Hu, X.; Amin, S.B.; Akdemir, K.C.; Seth, S.; Song, X.; Wang, Q.; et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017, 49, 349–357.
- Masutomi, K.; Possemato, R.; Wong, J.M.; Currier, J.L.; Tothova, Z.; Manola, J.B.; Ganesan, S.; Lansdorp, P.M.; Collins, K.; Hahn, W.C. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. USA 2005, 102, 8222–8227.
- Nitta, E.; Yamashita, M.; Hosokawa, K.; Xian, M.; Takubo, K.; Arai, F.; Nakada, S.; Suda, T. Telomerase reverse transcriptase protects ATM-deficient hematopoietic stem cells from ROS-induced apoptosis through a telomere-independent mechanism. Blood 2011, 117, 4169–4180.
- Koh, C.M.; Khattar, E.; Leow, S.C.; Liu, C.Y.; Muller, J.; Ang, W.X.; Li, Y.; Franzoso, G.; Li, S.; Guccione, E.; et al. Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J. Clin. Investig. 2015, 125, 2109–2122.
- Stewart, S.A.; Hahn, W.C.; O’Connor, B.F.; Banner, E.N.; Lundberg, A.S.; Modha, P.; Mizuno, H.; Brooks, M.W.; Fleming, M.; Zimonjic, D.B.; et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. USA 2002, 99, 12606–12611.
- Romaniuk, A.; Paszel-Jaworska, A.; Totoń, E.; Lisiak, N.; Hołysz, H.; Królak, A.; Grodecka-Gazdecka, S.; Rubiś, B. The non-canonical functions of telomerase: To turn off or not to turn off. Mol. Biol. Rep. 2019, 46, 1401–1411.
- Nassir, N.; Hyde, G.J.; Baskar, R. A telomerase with novel non-canonical roles: TERT controls cellular aggregation and tissue size in Dictyostelium. PLoS Genet. 2019, 15, e1008188.
- Saretzki, G. Extra-telomeric functions of human telomerase: Cancer, mitochondria and oxidative stress. Curr. Pharm. Des. 2014, 20, 6386–6403.
- Weinhold, N.; Jacobsen, A.; Schultz, N.; Sander, C.; Lee, W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 2014, 46, 1160–1165.
- Bell, R.J.; Rube, H.T.; Xavier-Magalhães, A.; Costa, B.M.; Mancini, A.; Song, J.S.; Costello, J.F. Understanding TERT Promoter Mutations: A Common Path to Immortality. Mol. Cancer Res. MCR 2016, 14, 315–323.
- Vinagre, J.; Almeida, A.; Pópulo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185.
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly recurrent TERT promoter mutations in human melanoma. Science 2013, 339, 957–959.
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K. TERT promoter mutations in familial and sporadic melanoma. Sciense 2013, 339, 959–961.
- Heidenreich, B.; Rachakonda, P.S.; Hemminki, K.; Kumar, R. TERT promoter mutations in cancer development. Curr. Opin. Genet. Dev. 2014, 24, 30–37.
- Sharma, S.; Chowdhury, S. Emerging mechanisms of telomerase reactivation in cancer. Trends Cancer 2022, 8, 632–641.
- Min, J.; Shay, J.W. TERT Promoter Mutations Enhance Telomerase Activation by Long-Range Chromatin Interactions. Cancer Discov. 2016, 6, 1212–1214.
- Bell, R.J.; Rube, H.T.; Kreig, A.; Mancini, A.; Fouse, S.D.; Nagarajan, R.P.; Choi, S.; Hong, C.; He, D.; Pekmezci, M.; et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 2015, 348, 1036–1039.
- Stern, J.L.; Theodorescu, D.; Vogelstein, B.; Papadopoulos, N.; Cech, T.R. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 2015, 29, 2219–2224.
- Borah, S.; Xi, L.; Zaug, A.J.; Powell, N.M.; Dancik, G.M.; Cohen, S.B.; Costello, J.C.; Theodorescu, D.; Cech, T.R. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 2015, 347, 1006–1010.
- Koelsche, C.; Sahm, F.; Capper, D.; Reuss, D.; Sturm, D.; Jones, D.T.; Kool, M.; Northcott, P.A.; Wiestler, B.; Böhmer, K.; et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013, 126, 907–915.
- Roh, M.R.; Park, K.H.; Chung, K.Y.; Shin, S.J.; Rha, S.Y.; Tsao, H. Telomerase reverse transcriptase (TERT) promoter mutations in Korean melanoma patients. Am. J. Cancer Res. 2017, 7, 134–138.
- Gandini, S.; Zanna, I.; De Angelis, S.; Palli, D.; Raimondi, S.; Ribero, S.; Masala, G.; Suppa, M.; Bellerba, F.; Corso, F.; et al. TERT promoter mutations and melanoma survival: A comprehensive literature review and meta-analysis. Crit. Rev. Oncol./Hematol. 2021, 160, 103288.
- Liu, X.; Wu, G.; Shan, Y.; Hartmann, C.; von Deimling, A.; Xing, M. Highly prevalent TERT promoter mutations in bladder cancer and glioblastoma. Cell Cycle 2013, 12, 1637–1638.
- Hurst, C.D.; Platt, F.M.; Knowles, M.A. Comprehensive mutation analysis of the TERT promoter in bladder cancer and detection of mutations in voided urine. Eur. Urol. 2014, 65, 367–369.
- Allory, Y.; Beukers, W.; Sagrera, A.; Flández, M.; Marqués, M.; Márquez, M.; van der Keur, K.A.; Dyrskjot, L.; Lurkin, I.; Vermeij, M.; et al. Telomerase reverse transcriptase promoter mutations in bladder cancer: High frequency across stages, detection in urine, and lack of association with outcome. Eur. Urol. 2014, 65, 360–366.
- Rachakonda, P.S.; Hosen, I.; de Verdier, P.J.; Fallah, M.; Heidenreich, B.; Ryk, C.; Wiklund, N.P.; Steineck, G.; Schadendorf, D.; Hemminki, K.; et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc. Natl. Acad. Sci. USA 2013, 110, 17426–17431.
- Wang, K.; Liu, T.; Ge, N.; Liu, L.; Yuan, X.; Liu, J.; Kong, F.; Wang, C.; Ren, H.; Yan, K.; et al. TERT promoter mutations are associated with distant metastases in upper tract urothelial carcinomas and serve as urinary biomarkers detected by a sensitive castPCR. Oncotarget 2014, 5, 12428–12439.
- Huang, D.S.; Wang, Z.; He, X.J.; Diplas, B.H.; Yang, R.; Killela, P.J.; Meng, Q.; Ye, Z.Y.; Wang, W.; Jiang, X.T.; et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur. J. Cancer 2015, 51, 969–976.
- Stoehr, R.; Taubert, H.; Zinnall, U.; Giedl, J.; Gaisa, N.T.; Burger, M.; Ruemmele, P.; Hurst, C.D.; Knowles, M.A.; Wullich, B.; et al. Frequency of TERT Promoter Mutations in Prostate Cancer. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 2015, 82, 53–57.
- Nonoguchi, N.; Ohta, T.; Oh, J.E.; Kim, Y.H.; Kleihues, P.; Ohgaki, H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013, 126, 931–937.
- Pezzuto, F.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Frequency and geographic distribution of TERT promoter mutations in primary hepatocellular carcinoma. Infect. Agents Cancer 2017, 12, 27.
- Lombardo, D.; Saitta, C.; Giosa, D.; Di Tocco, F.C.; Musolino, C.; Caminiti, G.; Chines, V.; Franzè, M.S.; Alibrandi, A.; Navarra, G.; et al. Frequency of somatic mutations in TERT promoter, TP53 and CTNNB1 genes in patients with hepatocellular carcinoma from Southern Italy. Oncol. Lett. 2020, 19, 2368–2374.
- Nault, J.C.; Mallet, M.; Pilati, C.; Calderaro, J.; Bioulac-Sage, P.; Laurent, C.; Laurent, A.; Cherqui, D.; Balabaud, C.; Zucman-Rossi, J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 2013, 4, 2218.
- Cevik, D.; Yildiz, G.; Ozturk, M. Common telomerase reverse transcriptase promoter mutations in hepatocellular carcinomas from different geographical locations. World J. Gastroenterol. 2015, 21, 311–317.
- Liu, X.; Bishop, J.; Shan, Y.; Pai, S.; Liu, D.; Murugan, A.K.; Sun, H.; El-Naggar, A.K.; Xing, M. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr.-Relat. Cancer 2013, 20, 603–610.
- Yang, H.; Park, H.; Ryu, H.J.; Heo, J.; Kim, J.S.; Oh, Y.L.; Choe, J.H.; Kim, J.H.; Kim, J.S.; Jang, H.W.; et al. Frequency of TERT Promoter Mutations in Real-World Analysis of 2,092 Thyroid Carcinoma Patients. Endocrinol. Metab. 2022, 37, 652–663.
- Alzahrani, A.S.; Alsaadi, R.; Murugan, A.K.; Sadiq, B.B. TERT Promoter Mutations in Thyroid Cancer. Horm. Cancer 2016, 7, 165–177.
- Campanella, N.C.; Celestino, R.; Pestana, A.; Scapulatempo-Neto, C.; de Oliveira, A.T.; Brito, M.J.; Gouveia, A.; Lopes, J.M.; Guimarães, D.P.; Soares, P.; et al. Low frequency of TERT promoter mutations in gastrointestinal stromal tumors (GISTs). Eur. J. Hum. Genet. EJHG 2015, 23, 877–879.
- Tallet, A.; Nault, J.C.; Renier, A.; Hysi, I.; Galateau-Sallé, F.; Cazes, A.; Copin, M.C.; Hofman, P.; Andujar, P.; Le Pimpec-Barthes, F.; et al. Overexpression and promoter mutation of the TERT gene in malignant pleural mesothelioma. Oncogene 2014, 33, 3748–3752.
- Griewank, K.G.; Schilling, B.; Murali, R.; Bielefeld, N.; Schwamborn, M.; Sucker, A.; Zimmer, L.; Hillen, U.; Schaller, J.; Brenn, T.; et al. TERT promoter mutations are frequent in atypical fibroxanthomas and pleomorphic dermal sarcomas. Mod. Pathol. 2014, 27, 502–508.
- Koelsche, C.; Renner, M.; Hartmann, W.; Brandt, R.; Lehner, B.; Waldburger, N.; Alldinger, I.; Schmitt, T.; Egerer, G.; Penzel, R.; et al. TERT promoter hotspot mutations are recurrent in myxoid liposarcomas but rare in other soft tissue sarcoma entities. J. Exp. Clin. Cancer Res. CR 2014, 33, 33.
- Scott, G.A.; Laughlin, T.S.; Rothberg, P.G. Mutations of the TERT promoter are common in basal cell carcinoma and squamous cell carcinoma. Mod. Pathol. 2014, 27, 516–523.
- Cheng, K.A.; Kurtis, B.; Babayeva, S.; Zhuge, J.; Tantchou, I.; Cai, D.; Lafaro, R.J.; Fallon, J.T.; Zhong, M. Heterogeneity of TERT promoter mutations status in squamous cell carcinomas of different anatomical sites. Ann. Diagn. Pathol. 2015, 19, 146–148.
- Zhao, Y.; Gao, Y.; Chen, Z.; Hu, X.; Zhou, F.; He, J. Low frequency of TERT promoter somatic mutation in 313 sporadic esophageal squamous cell carcinomas. Int. J. Cancer 2014, 134, 493–494.
- Kim, S.K.; Kim, J.H.; Han, J.H.; Cho, N.H.; Kim, S.J.; Kim, S.I.; Choo, S.H.; Kim, J.S.; Park, B.; Kwon, J.E. TERT promoter mutations in penile squamous cell carcinoma: High frequency in non-HPV-related type and association with favorable clinicopathologic features. J. Cancer Res. Clin. Oncol. 2021, 147, 1125–1135.
- Annunziata, C.; Pezzuto, F.; Greggi, S.; Ionna, F.; Losito, S.; Botti, G.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Distinct profiles of TERT promoter mutations and telomerase expression in head and neck cancer and cervical carcinoma. Int. J. Cancer 2018, 143, 1153–1161.
- Morris, L.G.T.; Chandramohan, R.; West, L.; Zehir, A.; Chakravarty, D.; Pfister, D.G.; Wong, R.J.; Lee, N.Y.; Sherman, E.J.; Baxi, S.S.; et al. The Molecular Landscape of Recurrent and Metastatic Head and Neck Cancers: Insights From a Precision Oncology Sequencing Platform. JAMA Oncol. 2017, 3, 244–255.
- Liu, R.; Xing, M. TERT promoter mutations in thyroid cancer. Endocr.-Relat. Cancer 2016, 23, R143–R155.
- Andrés-Lencina, J.J.; Rachakonda, S.; García-Casado, Z.; Srinivas, N.; Skorokhod, A.; Requena, C.; Soriano, V.; Kumar, R.; Nagore, E. TERT promoter mutation subtypes and survival in stage I and II melanoma patients. Int. J. Cancer 2019, 144, 1027–1036.
- Heidenreich, B.; Kumar, R. Altered TERT promoter and other genomic regulatory elements: Occurrence and impact. Int. J. Cancer 2017, 141, 867–876.
- Heidenreich, B.; Kumar, R. TERT promoter mutations in telomere biology. Mutat. Res. Rev. Mutat. Res. 2017, 771, 15–31.
More
Information
Subjects:
Oncology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
574
Revisions:
2 times
(View History)
Update Date:
07 Mar 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




