You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Marco Orlando | -- | 6219 | 2023-02-21 18:50:51 | | | |
| 2 | Marco Orlando | Meta information modification | 6219 | 2023-02-21 18:52:50 | | | | |
| 3 | Jessie Wu | Meta information modification | 6219 | 2023-02-23 08:40:09 | | | | |
| 4 | Jessie Wu | + 4 word(s) | 6223 | 2023-02-23 08:45:57 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Orlando, M.; Molla, G.; Castellani, P.; Pirillo, V.; Torretta, V.; Ferronato, N. Microbial Enzyme Applied to Plastic Depolymerization. Encyclopedia. Available online: https://encyclopedia.pub/entry/41495 (accessed on 25 December 2025).
Orlando M, Molla G, Castellani P, Pirillo V, Torretta V, Ferronato N. Microbial Enzyme Applied to Plastic Depolymerization. Encyclopedia. Available at: https://encyclopedia.pub/entry/41495. Accessed December 25, 2025.
Orlando, Marco, Gianluca Molla, Pietro Castellani, Valentina Pirillo, Vincenzo Torretta, Navarro Ferronato. "Microbial Enzyme Applied to Plastic Depolymerization" Encyclopedia, https://encyclopedia.pub/entry/41495 (accessed December 25, 2025).
Orlando, M., Molla, G., Castellani, P., Pirillo, V., Torretta, V., & Ferronato, N. (2023, February 21). Microbial Enzyme Applied to Plastic Depolymerization. In Encyclopedia. https://encyclopedia.pub/entry/41495
Orlando, Marco, et al. "Microbial Enzyme Applied to Plastic Depolymerization." Encyclopedia. Web. 21 February, 2023.
Copy Citation
The accumulation of synthetic plastic waste in the environment has become a global concern. Microbial enzymes (purified or as whole-cell biocatalysts) represent emerging biotechnological tools for waste circularity; they can depolymerize materials into reusable building blocks, but their contribution must be considered within the context of present waste management practices.
biotechnology
biodegradation
plastic
waste managment
circular economy
recycling
1. Introduction
Biotechnology tools have been employed for more than 20 years to depolymerize complex biomass waste into organic building blocks that, in turn, can be converted into useful intermediate feedstocks. These processes are typical applications of the circular (bio)economy concept. One of the most interesting examples is the conversion of lignocellulosic materials contained in plant waste from farming activities into bioethanol [1][2]. More recently, biotechnology strategies based on the use of microbial enzymes have been applied also to the degradation of petroleum-based plastics with the aim of providing greener alternatives to the traditional methods of recycling.
Various enzymes are used by bacterial or fungal organisms to fragment polymers into units of about 10–50 carbon atoms, which are used as carbon sources [3]. The catalytic capacity of highly purified stocks of enzymes (produced at an industrial level) can be exploited in biochemical processes performed under mild conditions (e.g., water solution in the 20–80 °C range) because they are biocatalysts evolved to act under conditions compatible with life. This allows for lowering of the economic and environmental costs of traditional processes, which usually require high temperatures and pressures [4]. The wide diversity in the metabolism of living organisms allowed for the discovery of biocatalysts suitable for many chemical reactions, including those responsible for plastic degradation, although consortia of whole organisms may also be required in some cases [5]. The enzymatic or microbial recycling of plastic materials is divided into three main steps: (i) the production of the biocatalysts, (ii) the depolymerization reaction of the polymer into monomers, and (iii) the purification and reuse of the monomers for resynthesis of the original material (closed-loop processes) or their bioconversion into different products (open-loop processes) [6].
A plethora of options is available for open-loop routes leveraging biotechnologies, as enzymes can catalyze multistep reactions to transform monomers into completely different molecules, which possibly have a value (per mass unit) superior to that of the original polymeric material [7]. The processes for open-loop plastic recycling can be performed in different bioreactors (employing purified enzymatic cocktails or whole organisms) or in a one-pot system [6]. The main classes of enzymes involved in plastic degradation and their products are reported in Figure 1. Table 1 reports the best-performing bio-based plastic depolymerization systems, which are predominantly implemented at laboratory scale.
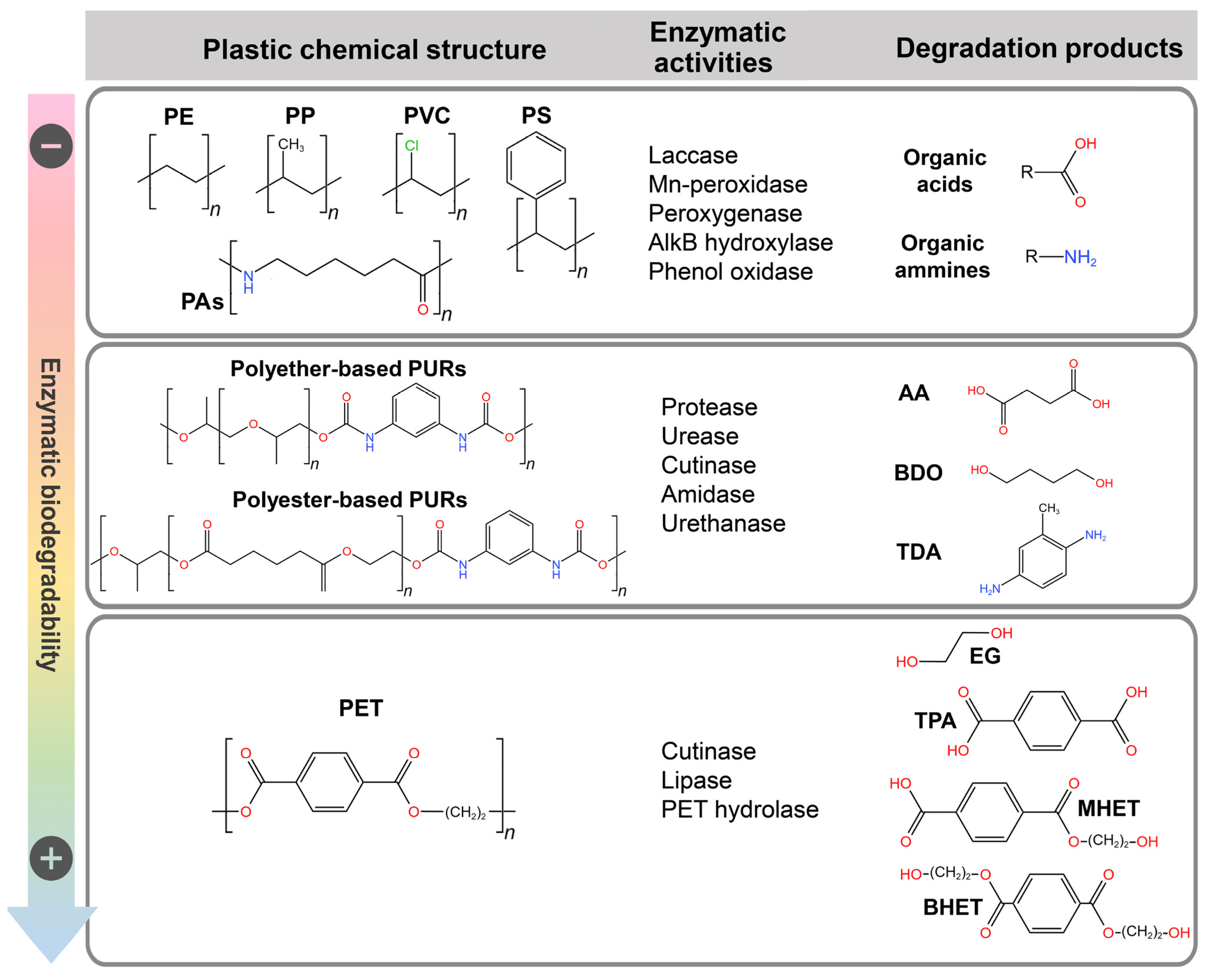
Figure 1. The main classes of enzymes involved in the degradation of different types of traditional petroleum-based plastics in order of their biodegradability to summarize what was reported in the text. Degradation products are exemplified. PURs—polyurethanes, AA—adipic acid, BDO—1,4-butanediol, TDA—toluene diamine, PE—polyethylene, PP—polypropylene, PAs—polyamides, PVC—polyvinylchloride, PS—polystyrene, PET—polyethylene terephthalate, EF—ethylene glycol, TPA—terephthalic acid, MHET—2-hydroxyethyl terephthalate, BHET—bis(2-hydroxyethyl) terephthalate.
Table 1. List of the most performant biological systems for traditional petroleum-based plastic degradation known to date. They are ordered by plastic polymer type and listed by decreasing degradation rate.
| Plastic Polymer | Material | Source | Reaction Condition | Degradation Rate | Reference |
|---|---|---|---|---|---|
| PET | Ultrathin PET film (2.5 to 7 nm) obtained from amorphous PET sheets of 2 mm thickness | IsPETaseTM [8] and IsMHETaseSM [9], two variants of I. sakaiensis PETase and METase |
PBS buffer, pH 7.4, 50 °C | ≈70% in 1 h | [10] |
| PET | Amorphized and micronized PET (≈200–250 µm) from post-consumer bottle-grade PET | Cutinase from leaf–branch compost (ICCG variant) |
100 mM potassium phosphate buffer, pH 8.0, 72 °C |
≈90% in 10 h | [11] |
| PET | 2 × 1 cm2 amorphous Goodfellow PET film and low-crystallinity (13%) PET powder | PHL7/PES-H1, a cutinase from a compost site (L92F/Q94Y variant) | 1 M potassium phosphate buffer, pH 8.0, 72 °C, shaking at 1000 rpm | ≈100% in 24 h | [12] |
| PET | 3 × 0.5 cm2 flakes of an amorphous PET clamshell container | PHL7/PES-H1, a cutinase from a compost site | 1 M potassium phosphate buffer, pH 8.0, 70 °C, shaking at 1000 rpm | >95% in 24 h | [13] |
| PET | Low-crystallinity (1.2 to 6.2%) discs (6 mm) from 51 different post-consumer PET products | FAST-PETase, a variant of I. sakaiensis PETase |
100 mM potassium phosphate buffer, pH 8.0, 50 °C, shaking at 180 rpm |
≈100% in 1 to 7 days | [14] |
| PET | PET/PE composite packaging tray lid (4 mg, thickness of 325 μm PET and 40 μm PE) | HotPETase, a directed-evolved variant of I. sakaiensis PETase |
pH 9.2, 50 mM gly-OH buffer with 4% BugBuster | ≈20% in 24 h | [15] |
| PET | ≈37% crystallinity PET microplastics (≈300 µm) | TS-ΔIsPET, a variant of PETase from Ideonella sakaiensis | 100 mM potassium phosphate buffer, pH 8.0, 40 °C, shaking | ≈26% in 2 days | [16] |
| Polyester-type PUR | Lab-prepared 10 µm thin PUR and segmented PUR urea films based on lysine diisocyanate | Papain, Bromelain, Ficin, Chymotrypsin, Proteinase K | PBS, pH 7.0, 37 °C | Up to ≈50% in 7 days | [17] |
| Polyester-type PUR | Impranil-DLN® | Pseudomonas putida A12 | pH 8.0, 25 °C |
92% in 4 days | [18] |
| Polyester-type PUR | Impranil-DLN® | Pestalotiopsis Microspora E2712A |
25 °C in a rotary incubator | 99% in 2 weeks | [19] |
| Polyether-type PUR | PUR foam | Tenebrio molitor | Guts of the larvae (probably microbiota-assisted) | 67% after 30 days | [20] |
| Polyester-type PUR | Lab-prepared 0.3 mm thin polycaprolactone thermoplastic PUR film (Capa 2302) | Amidase E4143 and esterase E3576 | PBS, pH 7.0, 37 °C | 33% in 51 days | [21] |
| Polyester-type PUR | Impranil-DLN® | Cladosporium pseudocladospo-rioides, Cladosporium tenuissimum, Cladosporium asperulatum, Cladosporium montecillanum, Aspergillus fumigatus, Penicillium chrysogenum |
25 °C, no shaking | 40–87% in 2 weeks |
[22] |
| Polyester-type PUR | Commercial 1 mm thin PUR film | Aspergillus flavus ITCC 6051 | 28 °C under 120 rpm shaking | 60.6% in 30 days | [23] |
| Polyester-type PUR | Lab-prepared ~0.2 mm thin PUR | Bacillus subtilis MZA-75 and Pseudomonas aeruginosa MZA-85 | 37 °C under 150 rpm shaking | 40% in 30 days | [24] |
| Polyether-type PUR | PUR foam used for commercial production of mattress cushioning | Cladosporium pseudocladospo-rioides, Cladosporium tenuissimum, Cladosporium asperulatum, Cladosporium montecillanum, Aspergillus fumigatus, Penicillium chrysogenum |
25–30 °C, no shaking | 10–65% in 21 days |
[22] |
| Polyether-type PUR | Cubical ether–PUR (1 cm3) | Alternaria sp. PURDK2 | 30 °C, no shaking | 27.5% in 70 days | [25] |
| LDPE | LDPE bag | Galleria mellonella | - | 13% in 14 h | [26][27][28] |
| LDPE | LDPE film | Pseudomonas citronellolis | 37 °C under 15 rpm shaking | 17.8% in 4 days | [29] |
| LDPE | LDPE powder | Cupriavidus necator H16 | 30 °C in a rotary incubator | 33.7% in 21 days | [30] |
| LDPE | LDPE foam | Tenebrio molitor | Gut of the larvae (probably microbiota-assisted) | 49.0% in 32 days | [31] |
| LDPE | 20 μm thin PE film | Oscillatoria subbrevis | - | 30% in 42 days | [32] |
| PS | PS pyrolysate oil | Pseudomonas putida CA-3 |
Four consecutive treatments at 30 °C for 48 h | 10% in 8 days | [33] |
| PS | Brominated high-impact PS | Bacillus sp. | 30 °C, no shaking | 23.7% in 30 days | [34] |
| PS | Brominated high-impact PS | Exiguobacterium sp. strain YT2 |
30 °C under 150 rpm shaking | 12.4% in 30 days | [35] |
| PP | Max 250 µm microplastic PP | Bacillus cereus | 33 °C under 150 rpm shaking | 12% in 40 days | [36] |
| PP | Isotactic PP strips | Pseudomonas sp., Vibrio sp., Aspergillus niger |
pH 7.0, 30 °C | 60% in 175 days | [37] |
2. Enzymatic Polyethylene Terephthalate Recycling: Potential Industrial-Scale Application
2.1. Enzymatic Degradation of Polyethylene Terephthalate
PET is a semicrystalline material made up of polymeric chains of alternating terephthalic acid (TPA) and ethylene glycol (EG) joined by ester bonds. In crystalline regions, the polymer chains are tightly packed in parallel, while in amorphous regions, the chains assume a disordered conformation. The degree of crystallinity depends on the production process and affects the chemophysical properties of the polymer which, in turn, are related to the final product performance. For instance, PET used in the production of bottles and textile fibers has a high crystallinity (between 30–40%), while PET is mainly amorphous in food packaging (crystallinity < 10%).
Research on biotechnological degradation of PET started in 2005 [38]. Since then, several microbial PET-hydrolyzing enzymes (PHEs) have been discovered for their ability to break down PET into mono-(2-hydroxyethyl)-terephthalate (MHET), bis-(2-hydroxyethyl)-terephthalate (BHET), TPA, and EG by hydrolyzing the ester bond [39] (Figure 1). Examples of the most active enzymes are reported in Table 1. These enzymes descend from the same ancestral carboxyl ester hydrolase of the GxSxG family [40]. They share the same catalytic mechanism and span different enzymatic classes specialized on different natural substrates: cutinase (EC 3.1.1.74), lipase (EC 3.1.1.3), and PETase (or PET hydrolase) (EC 3.1.1.101) [41][42][43]. Remarkably, the PETase class was created in 2016 after the discovery of a PHE from Ideonella sakaiensis (IsPETase), a bacterium isolated near a WWTP, with optimal PET degradation activity at 40 °C [44]. IsPETase and other I. sakaiensis enzymes were identified and shown to allow for the depolymerization and metabolic assimilation of PET degradation products as a carbon source [45]. The relevance of such a PET waste assimilation process in natural environments was questioned because the depolymerization of highly crystalline PET (such as that used to produce bottles) under natural conditions for the growth of the microorganism (30 to 40 °C) is highly inefficient [46][47]. Indeed, highly crystalline PET regions and low temperatures (<40 °C) greatly reduce the accessibility of PET chains to PHEs. Accordingly, the accessibility is maximal between 70 and 74 °C, near the glass transition temperature of bulk PET [48][49].
Consequently, known PHEs seem unsuitable for efficient degradation of untreated highly crystalline PET at room temperature [49][50]. This prompted a search for thermostable PHEs produced by protein engineering [8][11][12][14][51][52] or enzyme discovery [13][53][54][55]. Among such attempts, the company Carbios, in a public–private partnership with the Toulouse Biotechnology Institute and other global partners, was the first to produce a thermostabilized engineered variant of the leaf–branch compost cutinase (LC-cutinase) suitable for an industrial-scale recycling process of PET in 2020 [56]. Carbios built a demonstration plant in September 2021 with a business plan to extend the system to other companies under a paid license agreement by 2023 and revenues expected by 2025 [57].
The Carbios technology and other promising enzymatic PET recycling processes (Table 1) act on low-crystallinity submillimeter PET films [12][13][14][58] or amorphized/micronized post-consumer PET materials (C-ZYME®, based on the enzyme variants reported by Tournier et al. [11] and various patents (i.e., WO2017198786A1, EP3517608A1, and WO2020021118A1). In practice, post-consumer PET degradation processes were operated after sorting, washing, grinding, and amorphization. These pretreatment steps were already established for non-enzymatic recycling processes [59]. The step involving the use of enzymes was designed to be performed in <1 day in a buffered water suspension (pH between 7 and 9) inside a bioreactor containing a preprocessed polymer and close to the PET glass transition temperature. However, enzymatic incubation above 70 °C does not permit complete degradation of the amorphized PET due to an ageing process that results into polymer recrystallization after <10 h [60][61]. Recent studies [61][62][63] revealed relatively high degradation rates, even when performing the reaction at lower temperatures (50 to 60 °C), despite requiring more time (≥1 day). These milder conditions were also tested on untreated and non-micronized low-crystallinity post-consumer PET materials [13][14]. Moreover, Tarazona et al. recently demonstrated that a reduced PET thickness and an increased accessible surface area reduce the glass transition temperature by up to ≈40 °C (for nanometric film) [10]; while this can speed up PET degradation at lower temperatures and inhibit recrystallization, it does not block degradation-induced thermal inactivation, which consists of the accumulation of inactivated protein aggregates on the PET surface, causing the inhibition of further polymer degradation (a process called passivation).
Overall, promising industrial-scale PET degradation technologies require a buffered water suspension at ≥50 °C and PHEs engineered to be stable, efficient, and with low product inhibition (eventually by combining multiple enzymes) allowing for complete depolymerization as fast as possible in order to prevent PET recrystallization and passivation [10]. A recent work showed significant condition-dependent differences in the depolymerization process (buffer, pH, enzyme, and substrate loading) catalyzed by similar engineered PHE variants [62]. In addition, the enzymatic capacity to adsorb and turn-over during PET chain hydrolysis were recently observed to follow a tradeoff [64]. For these reasons, the biochemical kinetics and the mechanism of enzymatic PET degradation should be studied in detail to select the best enzymes and reaction conditions under which both the PHE activity is optimal and the PET polymer is most degradable, prior to scaling up the process [65]. Moreover, it is critical to set up a standard assay condition to compare different PHEs (different conditions are used for the enzymes reported in Table 1). As an example, a recently thermostabilized IsPETase (HotPETase) was shown to have initial degradation rates at 60 °C, much higher than the ICCG LC-cutinase used by Carbios on low- and high-crystalline materials (including a composite PET-PE film); however, it mainly produced MHET (TPA is the main product of ICCG LC-cutinase) and was inactivated after few hours on highly crystalline substrates, before the degradation was completed [15]. Furthermore, L92F/Q94Y PHL7 cutinase was more efficient than ICCG LC-cutinase on low-crystalline materials, while it was less active on highly crystalline PET powder [12]. Finally, different relative degradation rates on PET of different sizes and crystallinity were also measured when comparing many recently discovered PHEs [66], motivating the possible need for a cocktail of multiple PHEs for the degradation of PET materials of different crystallinity and subjected to different pretreatments.
2.2. Closed-Loop Polyethylene Terephthalate Recycling by Enzymatic Depolymerization
Carbios company demonstrated the feasibility of a closed-loop recycling process by showing that it is possible to use the monomers (TPA and EG) produced from PET depolymerization of post-consumer PET flakes to produce a PET bottle possessing the same properties as a bottle produced from virgin PET [11]. This process may also be fostered by monomers recovered from PET contained in textile polyester waste; an enzymatic cocktail of PHEs and other enzymes could be added to chemical treatments (in concert or in a sequential manner) to allow for separation and to reach a superior depolymerization yield of polymers without causing the high material losses associated with traditional methods. This is because the PET contained in fibers of commercialized textile products is often blended with other natural (i.e., cellulose and cotton) or synthetic (polyesters, nylons, elastane, etc.) polymers [67][68].
A recent work used Carbios data to conduct a technoeconomic analysis (TEA) of a closed-loop PET recycling process from bottle-derived post-consumer flakes [59]; a minimum production cost (MPC) for TPA of USD 1.93 kg−1 was estimated, most of which (>50%) was due to the price of petroleum-based PET flakes (feedstock PET) acquired from MRFs, the plastic pretreatment, and the recovery of the degradation products and co-products. In contrast, the step involving enzymes accounted only for 20% of the cost. Nevertheless, the model shows that besides the feedstock PET price, which is an external factor, the load of PET in the degradation solution and the rate of enzymatic conversion are the other dominant factors affecting MPC, enabling a reduction in cost of up to USD 1.60 kg−1 [59], which is less than double the average petroleum-based PET price in 2021 (up to USD 0.85 kg−1) [69]. It must be pointed out that this TEA estimated that the environmental impact of TPA obtained from the enzymatically based recycling process is nearly halved with respect to virgin TPA production. A recently published life cycle assessment (LCA) based on the USA economy confirmed the previous estimation of the production costs [59] but indicated that current enzymatic PET recycling technology exhibits a 1.2 to 17 times superior environmental impact relative to virgin polymer production [70]. NaOH usage, PET waste collection/shredding/amorphization, and electricity were estimated to be the highest-impact contributors. Sensitivity analysis indicated that, through interventions during all stages of the recycling process, from feedstock preparation to product recovery, it is possible to achieve an environmental impact similar or even lower than virgin polymer production. However, a superior enzymatic depolymerization efficiency without changing any other condition was estimated to contribute only up to 14%. Main contributions were estimated to be provided by minimization of PET losses during sorting, elimination of PET flake pretreatments, enzyme immobilization, and reaction buffer reuse. A major environmental benefit should originate from the reduction in NaOH consumption by using completely different buffers and PET loadings to facilitate TPA precipitation or by using more environment friendly (e.g., ammonia) or recoverable bases. Overall, this research suggested that further process optimization is required to minimize waste during feedstock preparation and product recovery.
Novel biocatalysts should allow for effective PET depolymerization under different buffer conditions (including organic solvents) of less pretreated PET-containing materials rather than showing a superior degradation rate under a broad range of pH and temperature conditions. Moreover, this process should be evaluated on different sources of PET waste, including those from commonly mixed products (i.e., textiles) for which traditional technologies are less effective and for which both the high crystallinity and the material substructure are expected to pose accessibility challenges to degradative enzymes [67]. The goal is the setup of a closed-loop enzyme-based process with a halved environmental impact compared to virgin polymers, compensating for the superior costs of PET produced from the enzymatic recycling process with an increased environmental gain.
2.3. Open-Loop Polyethylene Terephthalate Bio-Recycling
The use of biotechnological tools into open-loop processes could represent an opportunity to overcome the overpricing problem of closed-loop enzymatic PET recycling; enzymes and/or microorganisms can be used to assimilate and metabolically convert PET building blocks (TPA and EG) into chemically different intermediate bulk compounds or polymers with high added value [71]. This could also ensure the economic sustainability of treating low-quality plastics collected from contaminated environments. Several processes have been tested at the laboratory scale (Table 2).
Kim et al. [72] presented a two-organism system for the biological conversion of chemically depolymerized PET; TPA was converted into protocatechuic acid (PCA) and other PCA-derived compounds (i.e., gallic acid, pyrogallol, catechol, muconic acid, and vanillic acid) by an engineered E. coli strain, while G. oxydans KCCM 40109 was used to produce glycolic acid from EG. These compounds are important bulk chemicals to produce inks, bioplastics, flavorings, and fragrances. The same authors also demonstrated the possibility of using TPA produced by the enzymatic depolymerization of PET to produce catechol for the production of multifunctional coating materials [73]. Kang et al. [74] proposed the use of an engineered E. coli strain to convert TPA monomers from post-consumer PET into 2-pyrone-4,6-dicarboxylic acid, a valuable monomer for the synthesis of next-generation biodegradable plastics. A Pseudomonas putida KT2440 strain was engineered to use BHET (a different PET biodegradation product) to produce β-ketoadipic acid, a building block used to produce performance-advantaged nylons [75]. A one-pot system was proposed by Tiso et al. for the sequential enzymatic depolymerization of PET into EG and TPA [76], followed by Pseudomonas umsongensis GO16 conversion into intracellular polyhydroxyalkanoates (PHAs) and extracellular hydroxyalkanoyloxy alkanoate, which is used in a chemical copolymerization reaction to produce a novel bio-based poly(amide urethane) PUR. A similar system based on the use of an E. coli strain was proposed by Sadler et al. to produce vanillin flavor [77]. Liu et al. [78] proposed the first one-pot microbial recycling system from PET using two co-cultivated engineered organisms: a Yarrowia lipolytica Po1f strain expressing a PHE to depolymerize PET at low temperature and a Pseudomonas stutzeri strain to convert TPA into the bioplastic polyhydroxybutyrate. The same group showed that muconic acid can be produced with a single engineered Pseudomonas putida strain [79].
Table 2. List of recent open-loop microbial conversion attempts starting from PET and polyester-PUR degradation products. The different approaches are distinguished with respect to the processing strategies proposed in [6]. Abbreviations: EG—ethylene glycol, AA—adipic acid, BDO—1,4-butanediol, TPA—terephthalic acid, BHET—bis(2-hydroxyethyl) terephthalate, PHAs—polyhydroxyalkanoates, HAA—hydroxyalkanoyloxy alkanoate, PHBS—polyhydroxybutyrate.
| Approach | Enzymes or Microbial Strain(s) | Plastic Monomer | Conversion Products (Yields %) | Reference |
|---|---|---|---|---|
| Separate depolymerization and conversion | Pseudomonas putida strains JM37 and KT2440 | EG | Glyoxylic acid (≈20%) |
[80] |
| Three derived Pseudomonas putida KT2440 strains | AA, BDO, EG | Monorhamnolipids (<1%) |
[81] | |
| Escherichia coli strain | TPA, EG | Gallic acid (≈93%), pyrogallol (≈40%), muconic acid (≈85%), vanillic acid (≈40%), glycolic acid (≈98.6%) | [72] | |
| Escherichia coli strain | TPA | 2-pyrone-4,6-dicarboxylic acid (96.08%) | [74] | |
| Pseudomonas putida KT2440 strain | BHET | β-ketoadipic acid (≈76%) |
[75] | |
| Escherichia coli strain | TPA | Catechol (≈67%) | [73] | |
| Combined depolymerization and conversion | LCC 1, Pseudomonas umsongensis GO16 strain | TPA, EG | PHAs and HAA (<2%) |
[76] |
| LCC 2, Escherichia coli MG1655 RARE strain | TPA, EG | Vanillin (≈79%) | [77] | |
| Consolidated bioprocessing |
Yarrowia lipolytica Po1f expressing PETase 3, Pseudomonas stutzeri TPA3 | TPA, EG | PHB (≈2.5%) | [78] |
| Pseudomonas putida KT2440-tacRDL expressing LCC 4 | TPA, EG | Muconic acid (≈50%) | [79] |
These examples of open-loop PET recycling employing enzymes and microorganisms represent proof-of-principle experiments demonstrating the potential of microbial enzyme-driven upcycling processes.
3. Polyurethane Biodegradation Is Restricted to Polyester-Based Polymers
3.1. Enzymatic Degradation of Polyurethanes
PURs represent the largest fraction of plastic waste that is unrecyclable using traditional technologies. PUR polymers are formed by many different subtypes of two building blocks belonging to polyol (e.g., 1,4-butanediol, BDO) and isocyanate (e.g., toluene diamine, TDA) classes linked by urethane bonds (Figure 2). Moreover, isocyanates usually contain aromatic rings that increase the rigidity of the polymer and are covalently crosslinked to polyols of other chains by the formation of ester bonds in polyester-type PURs or by the formation of ether bonds in polyether-type PURs [83]. PURs are among the most diversified polymers, with different compositions in bonds, building blocks, and crosslinking distributions. This permits the generation of PUR materials with tailored physical properties [84]; however, the consequence is that a material-specific toolbox of enzymes with different activities is required for their degradation [85].
The enzymes identified to date to cause some degradation or weight loss of PURs are mainly promiscuous esterase (EC 3.1), urease (EC 3.5.1.5), protease (EC 3.4.21), and amidase (EC 3.5.1.4) enzymes [84]. Some esterases were shown to potentially degrade liquid dispersion (Impranil-DLN®) or bulk polyester-type PURs, but there is no evidence of their capacity to cleave urethane bonds [84]. Proteases and amidases have been shown to hydrolyze both urethane and ester bonds, but very low degradation rates were reported on commercial elastomer polyether-type PUR films or polyester-type PUR pellets [86]. A combined system involving an esterase and an amidase showed improved synergistic degradation (33%) after 51 days on a model PCL-based thermoplastic PUR [21]. A laccase-mediated (EC 1.10.3.2) system previously reported to degrade PE caused a reduction of a few percentage points in weight of lab synthesized polyether-type PURs in 18 days [87]. Urethanases (EC 3.5.1.75), which are enzymes capable of specifically hydrolyzing the urethane bond between polyols and isocyanate units, were recently discovered [88]; although their use for full depolymerization of PURs within a few days is very promising, these enzymes were used and demonstrated to be effective only on the urethane bonds in low-molecular-weight dicarbamate aromatic diamines obtained after the chemical depolymerization (by glycolysis) of non-urethane bonds in a polyether PUR foam. Therefore, this process is bio-based only in the last step.
Overall, even if enzymes capable of degrading the urethane and ester bonds in PURs have been discovered, processes that make exclusive use of enzymes have relatively high degradation rates only on synthetic or oligomeric PURs designed to be more biodegradable [89][90]. Therefore, the best options for PUR biodegradation are the use of environmental microbial strains or consortia, the design of more degradable novel PUR materials, or synergy with chemical methods of depolymerization [91].
3.2. Microbial Degradation of Polyurethanes
Some promising microbial degradation systems acting on polyester-type PURs have been tested at lab scale in the last 10 years. Impranil-DLN® was used as the sole carbon source by Pseudomonas putida A12, achieving 92% degradation within four days under mild conditions [18]. Bacillus subtilis MZA-75 and Pseudomonas aeruginosa MZA-85 were able to co-act on a polyester-type PUR film, resulting in 40% weight loss after 30 days [24]. Examples of polyester-type PUR biodegradation were also reported for fungal strains, both on Impranil-DLN® [19], a liquid varnish [22], and a commercial film [23]. Oligomeric fragments are usually reported as degradation products, which means that complete hydrolysis to single building blocks was not achieved. Moreover, microorganisms usually require other carbon sources to grow on PURs and may be susceptible to the accumulation of metabolically toxic plastic degradation products or additives, as observed for a Pseudomonas strain that released diamines from degraded PURs [81].
Polyether-type PURs show higher recalcitrance to microbial biodegradation, in particular when rigid aromatic polyisocyanate units are present, since they are less accessible and not susceptible to degradation by esterase activity [83]. Until now, only three studies have reported a significant biodegradation of polyether-type PUR materials: Tenebrio molitor larvae were shown to ingest polyether PUR foams efficiently, resulting in a significant mass loss of nearly 67% after 35 days. However, polyether PUR fragments were found in the frass of the larvae, indicating partial degradation [20]. Alternaria sp. strain PURDK2 caused 27.5% degradation of polyether-type PUR cubes after 10 weeks [25], and some Cladosporium fungal strains were reported to cause about 65% weight loss of a polyether-type PUR foam in 21 days [22].
Considering the reported degradation rates, the use of simple ad hoc synthesized or non-waste PUR products, and the lack of knowledge on the complex enzymatic machinery produced by microbial consortia [21], it is not currently possible to depict a practical industrial-scale biodegradation process, even for less recalcitrant polyester-type PUR waste. Moreover, solid polyester-type PURs and polyether-type PURs were only partially biodegraded (up to 50% at most), requiring a few weeks or even a few months. Therefore, for more recalcitrant PURs, a manly bio-based depolymerization process is still unfeasible.
In the next few years, it will be crucial to isolate the microbial enzymatic machinery involved in the degradation of less recalcitrant polyester PURs by using model PUR substrates and developing new specific assays (as presented by Liu et al. [92]). This will allow for mimicking of the whole-organism biodegradation action using an enzymatic cocktail or a stepwise addition of enzymes. Future investigations should also be focused on specific commercial PUR materials [84].
3.3. Closed-Loop and Open-Loop Polyurethane Bio-Recycling
The complex structure of PURs leads to a variety of degradation products, including organic acids, organic alcohols (e.g., EG), and diamines (Figure 1). Carboxylic acids and alcohols can be used for the synthesis of virgin PURs and other poly(ester–ether–urethane) materials [91]. Amines such as toluene diamine (TDA) can be recovered and used to directly synthesize polyamides or toluene isocyanate (TDI) to subsequently make virgin PURs (bio-based closed-loop recycling). Although the full depolymerization of a polyether-PUR foam was demonstrated with a chemoenzymatic approach [88], its application on a real waste PUR material and the use of the degradation products (polyether-polyols, diethylene glycol, and aromatic diamines) for the resynthesis of a waste-like PUR material was never attempted.
Some PUR degradation products have also been tested as microbial carbon sources (Table 2). EG can be used as a substrate to produce PHAs, which can be used to manufacture biodegradable medical devices, as well as glyoxylic acid, which is usually used for buffering solutions and other products to treat human skin and hair [80]. Recently, polyols (adipic acid (AA), 1,4-butanediol (BDO), and EG) were used as a carbon source for a mixed culture of three recombinant Pseudomonas putida KT2440 bacterial strains [81]. These bacteria can produce rhamnolipids, which are specialty biosurfactants applied in many industrial sectors. Coupling this process with an efficient extraction of toxic aromatic diamines contained in PURs (i.e., TDA), as proposed by Chen et al. [93], can be used to realize a green route for the open-loop bio-recycling of PUR degradation products.
4. Prospective for the Biological Degradation of Other Petroleum-Based Plastics
The biodegradation of other recalcitrant petroleum-based plastics (PAs and plastics with only C–C covalent bonds between their building blocks, i.e., PE, PS, PP, and PVC) is challenging; no degradative enzyme acting on the hydrolytic cleavage of the C-C bond is known. Moreover, some of these plastics (PE, PP) are completely crystalline, preventing enzymes from accessing the potential attack sites. Therefore, a multistep reaction catalyzed by a combination of abiotic pretreatments and different types of oxidative enzymes has been proposed [94].
There are many different types of PAs (polymers made of repeating units of aliphatic, semi-aromatic, or aromatic molecules linked via amide bonds), the most popular being nylon and Kevlar [95]. Since natural silk is also a PA from a chemical point of view, it was expected that enzymes able to degrade synthetic PAs do exist in nature. Unfortunately, to date, no known microorganism able to effectively degrade polymeric PAs has been identified, while it was demonstrated that several bacteria can act on short oligomers of linear or cyclic nylon and are possibly useful for recycling the byproducts of PA production [5][96]. A more recent study by Biundo et al. [97] demonstrated that it is also possible to engineer PHEs to abolish PET degrading activity and simultaneously increase the catalytic efficiency towards soluble PA oligomers. A manganese-dependent peroxidase from white rot fungus IZU-154 is the only enzyme reported to act on high-molecular-weight nylon fibers [98].
It was proposed that PE biodegradation only happens after abiotic (ultraviolet exposure) or enzymatic oxidation by different classes of oxidoreductase (alkane hydroxylase EC 1.14.15.3, laccase or multi-copper oxidase EC 1.10.3.2, peroxygenases EC 1.11.2, or Mn-peroxidase EC 1.11.1.13). This results in the introduction of aldehydic, ketonic, or alcoholic groups, which allows for subsequent processing by other enzymes. The latter can produce intermediate acid oligomers that are intracellularly taken-up by microorganisms and further metabolized [99][100]. Several microbial strains capable of degrading PE with this proposed system have been reported, including the gut microbiota of invertebrates [27][94]; animal phenol oxidase (EC 1.14.15.3) enzymes contained in the saliva of wax worm (Galleria mellonella) larvae were also demonstrated to be involved in the oxidative degradation of PE [28]. No system studied to date can achieve complete biodegradation without abiotic intervention. Photo-oxidative and thermal pretreatments, although expensive and technically demanding, were shown to oxidize and release oligomers from PE, improving their biodegradation rates [101]. In some cases, PE degradation of up to 30% was reported in one month [30][32], with even nearly complete bio-assimilation over >8 months [102]. In the absence of such pretreatments, the highest biodegradation rates, determined as polymer weight loss, were observed for LDPE films or bags (up to 10% within a few days to one month) [26][29][94]. These degradation rates were reported only for LDPE materials, while HDPE is less accessible to microorganism attack and much more recalcitrant [103].
Although PS is amorphous, its C–C backbone is more resistant to enzymatic cleavage than PE [6]. No enzyme has been identified with the ability to efficiently degrade high-molecular-weight PS polymers, despite some studies linking PS degradation to the same laccase and oxidoreductase enzymes associated with PE degradation [104]. Some organisms [34][35], including invertebrates assisted by their gut microbiome [105], were reported to cause significant weight loss in pretreated PS material, with rates of around 10% over one month. Another study reported that Pseudomonas putida CA-3 cultures can reach this degradation level in 8 days and produce PHAs. In this case PS was thermally pretreated at an elevated temperature (pyrolysis) to convert it to a styrene oil; this two-step process is very energetically demanding [33].
PP polymers can be present in three stereoisomeric forms, namely atactic, isotactic, and syndiotactic [106]. All PP forms are more resistant than PE to heat and to chemical attack, including the action of enzymes [101]. Some studies of PP biodegradation reported weight loss rates of about 10% per month. In all cases, these processes required a strong pretreatment with either γ- or UV-irradiation or heat or the use of polymer blends mixing PP and carbohydrates [36][37][107].
Finally, PVC, a synthetic polymer that contains chloride, is a form of waste that poses a serious pollution problem [108]. Very few studies have reported the biodegradation of low-molecular-weight PVC oligomers or film [101][109]. The capacity to degrade PVC was associated with the activity of catalase-peroxidase (EC 1.11.1.21) enzymes in white rot fungi [110] or from bacteria found in the gut of insects larvae [109]. Nevertheless, the use of microorganisms to remove PVC waste is still controversial because of the possible release of organochlorine compounds that may be highly toxic for the organisms themselves and the environment [101].
The fatty acids released by the oxidative bio-depolymerization of C-C plastics cannot be used in a closed-loop process of recycling, but they could be used as a carbon source for microbial growth to produce high-added-value products. While an open-loop fully bio-based process has not been established, in a recent proof-of-principle study, an open-loop recycling process was designed to produce β-ketoadipate or PHAs using microbial strains growing on organic acids obtained from chemically oxidative depolymerization of mixed resins or post-consumer waste (made with PS, HDPE, and PET) [111]; although promising, an LCA analysis is required to assess the industrial-scale feasibility of the proposed systems. A recent LCA evaluation focused on the use of fatty acids from chemically depolymerized and oxidized PE as a carbon source for growing a Pseudomonas putida KT2440 strain to target PHA production: the proposed approaches were more expensive or had a greater impact on the environment than waste-to-energy approaches [112].
5. Perspectives
European targets for plastic waste recycling rates are set by Directive 94/62/EC to 50% and 55% by 2025 and 2030, respectively. These targets are stricter for PPW, requiring that 65% and 70% of PPW be recycled by 2025 and 2030, respectively [113]. These targets can be achieved by summing the already recycled PCPW (≈20%) with the fraction (18,514 kt y−1, ≈50% PCPW) that researchers estimated to be potentially included in future recycling flows (Figure 2a). The polymer composition of this unrecycled waste plastic flow is reported in Figure 2b.
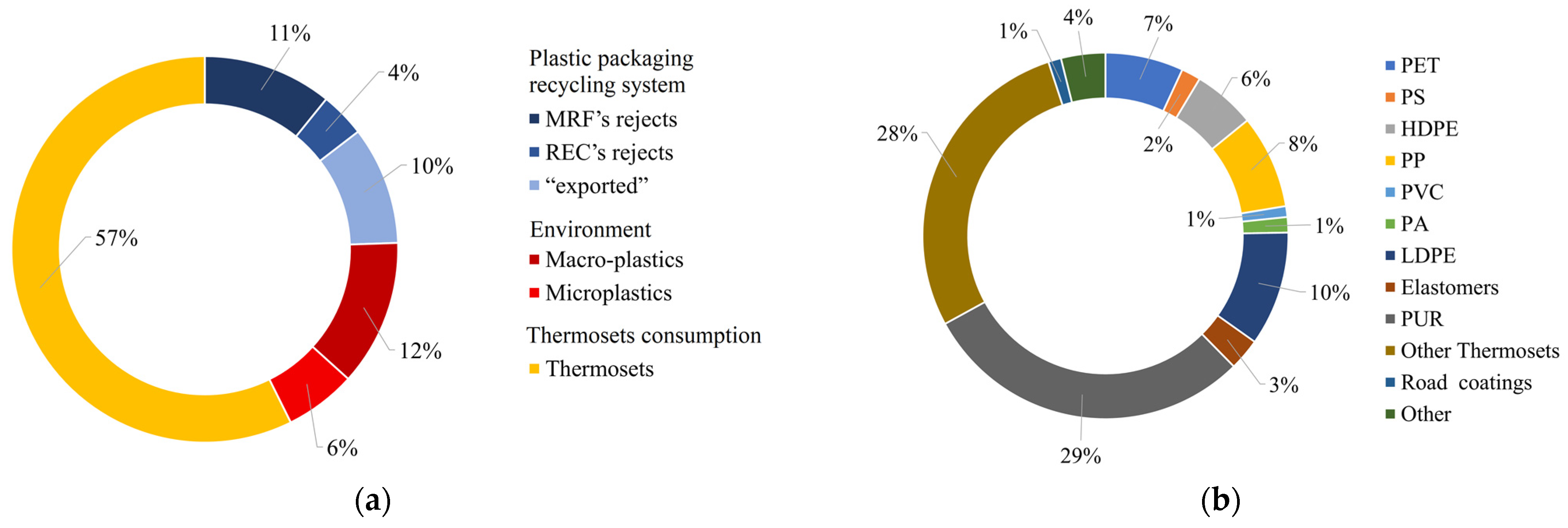
Figure 2. Unrecycled PCPW flows potentially included in future recyclable flows in the EU (Total 18,514 kt y−1): (a) divided by source; (b) divided by polymer composition. Elaborated from data collection and research conducted for this review article.The main contribution (57%) of this fraction is thermoset plastics (29.4% PURs and 27.9% other thermosets) (Figure 2a). Therefore, the most urgent interventions should focus on finding alternative recycling options to allow for the production of virgin thermoset materials from waste.
Thermoplastic rejects from MRFs or RECs account for 25% of this unrecycled PCPW flow (Figure 2a). Exported plastic waste (10%) was included, as the European Commission has stated that all PPW should be recycled in the EU market by 2030 after the Chinese government’s ban (April 2017) on imported recyclable solid waste [114]. An improvement in the amount of recycled plastic should be derived from improved sorting and preprocessing to reach higher purity of PCPW fractions at RECs; this is achievable by coupling traditional sorting technologies (i.e., manual sorting and NIR sensors) with machine-learning-based recognition of plastic waste from images [115][116]. This is expected to only increase the recycling rate in RECs, while thermoplastics rejects generated by mixed and small-particle plastics would remain an issue. This suggests that a fundamental change in the plastic value chain is required for marketable plastic materials to account for end-of-life circularity options in waste treatment processes, especially during the sorting step.
Although environmentally dispersed plastics are reported to be the smallest flow of unrecycled PCPW (18%, Figure 2a), they also represent the most complicated fraction to be recaptured (microplastics) and valorized (the quality of macroplastics recovered from the environment is poor). This flow should be lowered as much as possible by improving plastic collection rates through a focus on public behavior and municipal policies aimed at decreasing environmental leakages and progressively increasing the amount of collected plastics to target recycling.
The remining PCPW mixed with non-plastic materials and disposed of as non-plastic unrecyclable waste (targeted to landfilling, incineration, or recovery) was not considered as a potentially recyclable flow.
Considering the necessary interventions, microbial enzyme-based biotechnology could be integrated in the waste management sector and contribute to the introduction of new depolymerization methods for thermosets and rejected thermoplastics, allowing for resynthesis of virgin polymers or the open-loop upcycling of degradation products. Therefore, it is necessary to make biotechnological solutions available that are suitable to treat plastic waste of different quality, including materials made of (or blended with) many different polymers (not necessarily synthetic) that are difficult to separate with traditional methods prior to recycling; moreover, such systems should retain market competitiveness and an environmental impact equal to or lower than that of existing technologies. In this regard, the actual contribution of biotechnology to plastic degradation is limited, as only the bio-based depolymerization systems described for amorphous aliphatic polyesters (such as pretreated PET) and liquid polyester-type PURs have the potential to be exploited at the industrial scale, although not competitive with traditional approaches at present. It will be crucial to improve such systems by further enzyme discovery or engineering studies to increase the degradation efficiency (to overcome the passivation issue) under high PET load or for less pretreated materials.
Even if PET is, by far, the most consumed polyester, PET waste represents only a minor fraction (≈7%) of unrecycled flows with respect to PURs, thermosets, and other non-polyester plastics (Figure 2b). A synergy with promising methods of abiotic secondary and tertiary PUR depolymerization approaches could allow for depolymerization of recalcitrant PUR waste, but no method has been tested on real PUR waste to date [88][91][117][118][119]. PAs and C-C backbone plastics, such as PE and PS, are currently much less efficiently biodegraded. Moreover, some of these plastics (i.e., PVC) may release toxic components after their degradation. Although biodegradation could be enhanced by new methods of abiotic preprocessing in the presence of organometallic catalysts, these treatments require a lot of energy and relatively high temperatures (>150 °C), thus contributing to higher costs and environmental impact [6][120]. Moreover, as most biodegradation processes have only been conducted at laboratory scale [99][101], without biochemical information on the kinetics of the involved reactions, with scarce data on degradation products, and often observing that mainly additives were cleaved [121], a fully bio-based depolymerization of non-polyester petroleum-based plastics is predicted not to be economically and environmentally competitive with traditional approaches within a 10-year time frame. Instead, it is necessary to look at new approaches of tertiary recycling [122][123][124] and to use them in combination with microbial enzymes to convert degradation products into high-added-value chemicals [111][125][126].
The difficulties to apply enzymes for depolymerizing non-polyester plastic waste fractions indicate that particular attention should be taken on aliphatic polyesters (i.e., PEF, PBAT, and PBS) to replace traditional recalcitrant petroleum-based plastic waste. Regardless of whether these plastics are biodegradable or bio-based, researchers argue that the most important aspect is the creation of suitable technologies to allow for their bio-based degradation and circular production from waste within an industrial setup, while avoiding environmental leakages.
In conclusion, the use of enzymatic microbial biotechnologies for plastic degradation and valorization requires a rethink of the whole plastic value chain, as well as the development and marketing of alternative plastic materials (made with non-food biomass derivatives or aliphatic polyesters that have a biodegradability similar or superior to that of PET), the redesign of plastic packaging and products, the improvement of collection and sorting methods, and their tight integration with new, eventually chemo-assisted bio-based systems of degradation and recycling (competitive from an economical point of view and with a lower environmental impact compared to available technologies). This, in turn, will require stronger co-operation between biotechnologists and stakeholders involved with feedstock resources, material production, and waste management to find novel effective industrial technologies and regulations for a transition to a circular (bio)economy in plastic waste management.
References
- Yang, S.; Hai, F.I.; Nghiem, L.D.; Price, W.E.; Roddick, F.; Moreira, M.T.; Magram, S.F. Understanding the factors controlling the removal of trace organic contaminants by white-rot fungi and their lignin modifying enzymes: A critical review. Bioresour. Technol. 2013, 141, 97–108.
- Park, G.W.; Gong, G.; Joo, J.C.; Song, J.; Lee, J.; Lee, J.-P.; Kim, H.T.; Ryu, M.H.; Sirohi, R.; Zhuang, X.; et al. Recent progress and challenges in biological degradation and biotechnological valorization of lignin as an emerging source of bioenergy: A state-of-the-art review. Renew. Sustain. Energy Rev. 2022, 157, 112025.
- Antranikian, G.; Streit, W.R. Microorganisms harbor keys to a circular bioeconomy making them useful tools in fighting plastic pollution and rising CO2 levels. Extremophiles 2022, 26, 10.
- Abdelraheem, E.M.; Busch, H.; Hanefeld, U.; Tonin, F. Biocatalysis explained: From pharmaceutical to bulk chemical production. React. Chem. Eng. 2019, 4, 1878–1894.
- Danso, D.; Chow, J.; Streit, W.R. Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019, 85, e01095-19.
- Ellis, L.D.; Rorrer, N.A.; Sullivan, K.P.; Otto, M.; McGeehan, J.E.; Román-Leshkov, Y.; Wierckx, N.; Beckham, G.T. Chemical and biological catalysis for plastics recycling and upcycling. Nat. Catal. 2021, 4, 539–556.
- Lee, S.; Lee, Y.R.; Kim, S.J.; Lee, J.-S.; Min, K. Recent advances and challenges in the biotechnological upcycling of plastic wastes for constructing a circular bioeconomy. Chem. Eng. J. 2022, 454, 140470.
- Brott, S.; Pfaff, L.; Schuricht, J.; Schwarz, J.N.; Böttcher, D.; Badenhorst, C.P.; Wei, R.; Bornscheuer, U.T. Engineering and evaluation of thermostable IsPETase variants for PET degradation. Eng. Life Sci. 2022, 22, 192–203.
- Palm, G.J.; Reisky, L.; Böttcher, D.; Müller, H.; Michels, E.A.; Walczak, M.C.; Berndt, L.; Weiss, M.S.; Bornscheuer, U.T.; Weber, G.; et al. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nat. Commun. 2019, 10, 1717.
- Tarazona, N.A.; Wei, R.; Brott, S.; Pfaff, L.; Bornscheuer, U.T.; Lendlein, A.; Machatschek, R. Rapid depolymerization of poly (ethylene terephthalate) thin films by a dual-enzyme system and its impact on material properties. Chem Catal. 2022, 2, 3573–3589.
- Tournier, V.; Topham, C.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.-L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219.
- Pfaff, L.; Gao, J.; Li, Z.; Jäckering, A.; Weber, G.; Mican, J.; Chen, Y.; Dong, W.; Han, X.; Feiler, C.G.; et al. Multiple substrate binding mode-guided engineering of a thermophilic PET hydrolase. ACS Catal. 2022, 12, 9790–9800.
- Sonnendecker, C.; Oeser, J.; Richter, P.K.; Hille, P.; Zhao, Z.; Fischer, C.; Lippold, H.; Blázquez-Sánchez, P.; Engelberger, F.; Ramírez-Sarmiento, C.A.; et al. Low Carbon Footprint Recycling of Post-Consumer PET Plastic with a Metagenomic Polyester Hydrolase. ChemSusChem 2022, 15, e202101062.
- Lu, H.; Diaz, D.J.; Czarnecki, N.J.; Zhu, C.; Kim, W.; Shroff, R.; Acosta, D.J.; Alexander, B.R.; Cole, H.O.; Zhang, Y.; et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 2022, 604, 662–667.
- Bell, E.L.; Smithson, R.; Kilbride, S.; Foster, J.; Hardy, F.J.; Ramachandran, S.; Tedstone, A.A.; Haigh, S.J.; Garforth, A.A.; Day, P.J.; et al. Directed evolution of an efficient and thermostable PET depolymerase. Nat. Catal. 2022, 5, 673–681.
- Pirillo, V.; Orlando, M.; Tessaro, D.; Pollegioni, L.; Molla, G. An efficient protein evolution workflow for the improvement of bacterial PET hydrolyzing enzymes. Int. J. Mol. Sci. 2022, 23, 264.
- Yamamoto, N.; Nakayama, A.; Oshima, M.; Kawasaki, N.; Aiba, S.-I. Enzymatic hydrolysis of lysine diisocyanate based polyurethanes and segmented polyurethane ureas by various proteases. React. Funct. Polym. 2007, 67, 1338–1345.
- Peng, Y.-H.; Shih, Y.-H.; Lai, Y.-C.; Liu, Y.-Z.; Liu, Y.-T.; Lin, N.-C. Degradation of polyurethane by bacterium isolated from soil and assessment of polyurethanolytic activity of a Pseudomonas putida strain. Environ. Sci. Pollut. Res. 2014, 21, 9529–9537.
- Russell, J.R.; Huang, J.; Anand, P.; Kucera, K.; Sandoval, A.G.; Dantzler, K.W.; Hickman, D.; Jee, J.; Kimovec, F.M.; Koppstein, D.; et al. Biodegradation of polyester polyurethane by endophytic fungi. Appl. Environ. Microbiol. 2011, 77, 6076–6084.
- Liu, J.; Liu, J.; Xu, B.; Xu, A.; Cao, S.; Wei, R.; Zhou, J.; Jiang, M.; Dong, W. Biodegradation of polyether-polyurethane foam in yellow mealworms (Tenebrio molitor) and effects on the gut microbiome. Chemosphere 2022, 304, 135263.
- Magnin, A.; Pollet, E.; Perrin, R.; Ullmann, C.; Persillon, C.; Phalip, V.; Avérous, L. Enzymatic recycling of thermoplastic polyurethanes: Synergistic effect of an esterase and an amidase and recovery of building blocks. Waste Manag. 2019, 85, 141–150.
- Álvarez-Barragán, J.; Domínguez-Malfavón, L.; Vargas-Suárez, M.; González-Hernández, R.; Aguilar-Osorio, G.; Loza-Tavera, H. Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams. Appl. Environ. Microbiol. 2016, 82, 5225–5235.
- Mathur, G.; Prasad, R. Degradation of polyurethane by Aspergillus flavus (ITCC 6051) isolated from soil. Appl. Biochem. Biotechnol. 2012, 167, 1595–1602.
- Shah, Z.; Gulzar, M.; Hasan, F.; Shah, A.A. Degradation of polyester polyurethane by an indigenously developed consortium of Pseudomonas and Bacillus species isolated from soil. Polym. Degrad. Stab. 2016, 134, 349–356.
- Matsumiya, Y.; Murata, N.; Tanabe, E.; Kubota, K.; Kubo, M. Isolation and characterization of an ether-type polyurethane-degrading micro-organism and analysis of degradation mechanism by Alternaria sp. J. Appl. Microbiol. 2010, 108, 1946–1953.
- Bombelli, P.; Howe, C.J.; Bertocchini, F. Polyethylene bio-degradation by caterpillars of the wax moth Galleria mellonella. Curr. Biol. 2017, 27, R292–R293.
- Barrionuevo, J.M.R.; Vilanova-Cuevas, B.; Alvarez, A.; Martín, E.; Malizia, A.; Galindo-Cardona, A.; de Cristóbal, R.E.; Occhionero, M.A.; Chalup, A.; Monmany-Garzia, A.C.; et al. The bacterial and fungal gut microbiota of the greater wax moth, Galleria mellonella L. consuming polyethylene and polystyrene. Front. Microbiol. 2022, 13, 918861.
- Sanluis-Verdes, A.; Colomer-Vidal, P.; Rodriguez-Ventura, F.; Bello-Villarino, M.; Spinola-Amilibia, M.; Ruiz-Lopez, E.; Illanes-Vicioso, R.; Castroviejo, P.; Cigliano, R.A.; Montoya, M.; et al. Wax worm saliva and the enzymes therein are the key to polyethylene degradation by Galleria mellonella. Nat. Commun. 2022, 13, 5568.
- Bhatia, M.; Girdhar, A.; Tiwari, A.; Nayarisseri, A. Implications of a novel Pseudomonas species on low density polyethylene biodegradation: An in vitro to in silico approach. SpringerPlus 2014, 3, 497.
- Montazer, Z.; Habibi Najafi, M.B.; Levin, D.B. Microbial degradation of low-density polyethylene and synthesis of polyhydroxyalkanoate polymers. Can. J. Microbiol. 2019, 65, 224–234.
- Brandon, A.M.; Gao, S.-H.; Tian, R.; Ning, D.; Yang, S.-S.; Zhou, J.; Wu, W.-M.; Criddle, C.S. Biodegradation of polyethylene and plastic mixtures in mealworms (larvae of Tenebrio molitor) and effects on the gut microbiome. Environ. Sci. Technol. 2018, 52, 6526–6533.
- Sarmah, P.; Rout, J. Efficient biodegradation of low-density polyethylene by cyanobacteria isolated from submerged polyethylene surface in domestic sewage water. Environ. Sci. Pollut. Res. 2018, 25, 33508–33520.
- Ward, P.G.; Goff, M.; Donner, M.; Kaminsky, W.; O’Connor, K.E. A two step chemo-biotechnological conversion of polystyrene to a biodegradable thermoplastic. Environ. Sci. Technol. 2006, 40, 2433–2437.
- Mohan, A.J.; Sekhar, V.C.; Bhaskar, T.; Nampoothiri, K.M. Microbial assisted high impact polystyrene (HIPS) degradation. Bioresour. Technol. 2016, 213, 204–207.
- Sekhar, V.C.; Nampoothiri, K.M.; Mohan, A.J.; Nair, N.R.; Bhaskar, T.; Pandey, A. Microbial degradation of high impact polystyrene (HIPS), an e-plastic with decabromodiphenyl oxide and antimony trioxide. J. Hazard. Mater. 2016, 318, 347–354.
- Auta, H.; Emenike, C.; Fauziah, S. Screening of Bacillus strains isolated from mangrove ecosystems in Peninsular Malaysia for microplastic degradation. Environ. Pollut. 2017, 231, 1552–1559.
- Cacciari, I.; Quatrini, P.; Zirletta, G.; Mincione, E.; Vinciguerra, V.; Lupattelli, P.; Giovannozzi Sermanni, G. Isotactic polypropylene biodegradation by a microbial community: Physicochemical characterization of metabolites produced. Appl. Environ. Microbiol. 1993, 59, 3695–3700.
- Müller, R.J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.D. Enzymatic degradation of poly (ethylene terephthalate): Rapid hydrolyse using a hydrolase from T. fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405.
- Webb, H.K.; Arnott, J.; Crawford, R.J.; Ivanova, E.P. Plastic degradation and its environmental implications with special reference to poly (ethylene terephthalate). Polymers 2012, 5, 1–18.
- Oh, C.; Kim, T.D.; Kim, K.K. Carboxylic ester hydrolases in bacteria: Active site, structure, function and application. Crystals 2019, 9, 597.
- Danso, D.; Schmeisser, C.; Chow, J.; Zimmermann, W.; Wei, R.; Leggewie, C.; Li, X.; Hazen, T.; Streit, W.R. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl. Environ. Microbiol. 2018, 84, e02773-17.
- Carr, C.M.; Clarke, D.J.; Dobson, A.D. Microbial polyethylene terephthalate hydrolases: Current and future perspectives. Front. Microbiol. 2020, 11, 2825.
- Kawai, F. The current state of research on PET hydrolyzing enzymes available for biorecycling. Catalysts 2021, 11, 206.
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K.; et al. A bacterium that degrades and assimilates poly (ethylene terephthalate). Science 2016, 351, 1196–1199.
- Yoshida, S.; Hiraga, K.; Taniguchi, I.; Oda, K. Ideonella sakaiensis, PETase, and MHETase: From identification of microbial PET degradation to enzyme characterization. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 648, pp. 187–205.
- Yang, Y.; Yang, J.; Jiang, L. Comment on “A bacterium that degrades and assimilates poly (ethylene terephthalate)”. Science 2016, 353, 759.
- Wallace, N.E.; Adams, M.C.; Chafin, A.C.; Jones, D.D.; Tsui, C.L.; Gruber, T.D. The highly crystalline PET found in plastic water bottles does not support the growth of the PETase-producing bacterium Ideonella sakaiensis. Environ. Microbiol. Rep. 2020, 12, 578–582.
- Falkenstein, P.; Wei, R.; Matysik, J.; Song, C. Mechanistic investigation of enzymatic degradation of polyethylene terephthalate by nuclear magnetic resonance. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 648, pp. 231–252.
- Brizendine, R.K.; Erickson, E.; Haugen, S.J.; Ramirez, K.J.; Miscall, J.; Salvachúa, D.; Pickford, A.R.; Sobkowicz, M.J.; McGeehan, J.E.; Beckham, G.T.; et al. Particle size reduction of poly (ethylene terephthalate) increases the rate of enzymatic depolymerization but does not increase the overall conversion extent. ACS Sustain. Chem. Eng. 2022, 10, 9131–9140.
- Thomsen, T.B.; Hunt, C.J.; Meyer, A.S. Influence of substrate crystallinity and glass transition temperature on enzymatic degradation of polyethylene terephthalate (PET). New Biotechnol. 2022, 69, 28–35.
- Son, H.F.; Joo, S.; Seo, H.; Sagong, H.-Y.; Lee, S.H.; Hong, H.; Kim, K.-J. Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzym. Microb. Technol. 2020, 141, 109656.
- Cui, Y.; Chen, Y.; Liu, X.; Dong, S.; Tian, Y.E.; Qiao, Y.; Mitra, R.; Han, J.; Li, C.; Han, X. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy. ACS Catal. 2021, 11, 1340–1350.
- Meyer-Cifuentes, I.E.; Werner, J.; Jehmlich, N.; Will, S.E.; Neumann-Schaal, M.; Öztürk, B. Synergistic biodegradation of aromatic-aliphatic copolyester plastic by a marine microbial consortium. Nat. Commun. 2020, 11, 5790.
- Xi, X.; Ni, K.; Hao, H.; Shang, Y.; Zhao, B.; Qian, Z. Secretory expression in Bacillus subtilis and biochemical characterization of a highly thermostable polyethylene terephthalate hydrolase from bacterium HR29. Enzym. Microb. Technol. 2021, 143, 109715.
- Eiamthong, B.; Meesawat, P.; Wongsatit, T.; Jitdee, J.; Sangsri, R.; Patchsung, M.; Aphicho, K.; Suraritdechachai, S.; Huguenin-Dezot, N.; Tang, S.; et al. Discovery and genetic code expansion of a polyethylene terephthalate (PET) hydrolase from the human saliva metagenome for the degradation and bio-functionalization of PET. Angew. Chem. 2022, 61, e202203061.
- Sulaiman, S.; Yamato, S.; Kanaya, E.; Kim, J.-J.; Koga, Y.; Takano, K.; Kanaya, S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 2012, 78, 1556–1562.
- Carbios—Enzymes Powering the Circular Economy. Available online: https://www.carbios.com/wp-content/uploads/2022/01/carbios_presentation-january-2022.pdf (accessed on 20 October 2022).
- García, J.L. Enzymatic recycling of polyethylene terephthalate through the lens of proprietary processes. Microb. Biotechnol. 2022, 15, 2699–2704.
- Singh, A.; Rorrer, N.A.; Nicholson, S.R.; Erickson, E.; DesVeaux, J.S.; Avelino, A.F.; Lamers, P.; Bhatt, A.; Zhang, Y.; Avery, G.; et al. Techno-economic, life-cycle, and socioeconomic impact analysis of enzymatic recycling of poly (ethylene terephthalate). Joule 2021, 5, 2479–2503.
- Pasula, R.R.; Lim, S.; Ghadessy, F.J.; Sana, B. The influences of substrates’ physical properties on enzymatic PET hydrolysis: Implications for PET hydrolase engineering. Eng. Biol. 2022, 6, 17–22.
- Wei, R.; von Haugwitz, G.; Pfaff, L.; Mican, J.; Badenhorst, C.P.; Liu, W.; Weber, G.; Austin, H.P.; Bednar, D.; Damborsky, J.; et al. Mechanism-based design of efficient PET hydrolases. ACS Catal. 2022, 12, 3382–3396.
- Erickson, E.; Shakespeare, T.J.; Bratti, F.; Buss, B.L.; Graham, R.; Hawkins, M.A.; König, G.; Michener, W.E.; Miscall, J.; Ramirez, K.J.; et al. Comparative performance of PETase as a function of reaction conditions, substrate properties, and product accumulation. ChemSusChem 2022, 15, e202101932.
- Pirillo, V.; Orlando, M.; Battaglia, C.; Pollegioni, L.; Molla, G. Efficient polyethylene terephthalate degradation at moderate temperature: A protein engineering study of LC-cutinase highlights the key role of residue 243. FEBS J. 2023, accepted.
- Bååth, J.A.; Jensen, K.; Borch, K.; Westh, P.; Kari, J. Sabatier principle for rationalizing enzymatic hydrolysis of a synthetic polyester. JACS Au 2022, 2, 1223–1231.
- Schubert, S.W.; Schaller, K.; Bååth, J.A.; Hunt, C.; Borch, K.; Jensen, K.; Brask, J.; Westh, P. Reaction pathways for the enzymatic degradation of poly (ethylene terephthalate): What characterizes an efficient PET-hydrolase? ChemBioChem 2022, 24, e202200516.
- Erickson, E.; Gado, J.E.; Avilán, L.; Bratti, F.; Brizendine, R.K.; Cox, P.A.; Gill, R.; Graham, R.; Kim, D.-J.; König, G.; et al. Sourcing thermotolerant poly (ethylene terephthalate) hydrolase scaffolds from natural diversity. Nat. Commun. 2022, 13, 7850.
- Jönsson, C.; Wei, R.; Biundo, A.; Landberg, J.; Schwarz Bour, L.; Pezzotti, F.; Toca, A.; Jacques, L.M.; Bornscheuer, U.T.; Syrén, P.O. Biocatalysis in the recycling landscape for synthetic polymers and plastics towards circular textiles. ChemSusChem 2021, 14, 4028–4040.
- Quartinello, F.; Vajnhandl, S.; Volmajer Valh, J.; Farmer, T.J.; Vončina, B.; Lobnik, A.; Herrero Acero, E.; Pellis, A.; Guebitz, G.M. Synergistic chemo-enzymatic hydrolysis of poly (ethylene terephthalate) from textile waste. Microb. Biotechnol. 2017, 10, 1376–1383.
- Statista. Price of Polyethylene Terephthalate (PET) Worldwide from 2017 to 2020 with Estimated Figures for 2021 to 2022. 2022. Available online: https://www.statista.com/statistics/1171088/price-polyethylene-terephthalate-forecast-globally/ (accessed on 30 May 2022).
- Uekert, T.; DesVeaux, J.S.; Singh, A.; Nicholson, S.R.; Lamers, P.; Ghosh, T.; McGeehan, J.E.; Carpenter, A.C.; Beckham, G.T. Life cycle assessment of enzymatic poly (ethylene terephthalate) recycling. Green Chem. 2022, 24, 6531–6543.
- Lee, A.; Liew, M.S. Tertiary recycling of plastics waste: An analysis of feedstock, chemical and biological degradation methods. J. Mater. Cycles Waste Manag. 2021, 23, 32–43.
- Kim, H.T.; Kim, J.K.; Cha, H.G.; Kang, M.J.; Lee, H.S.; Khang, T.U.; Yun, E.J.; Lee, D.-H.; Song, B.K.; Park, S.J.; et al. Biological valorization of poly (ethylene terephthalate) monomers for upcycling waste PET. ACS Sustain. Chem. Eng. 2019, 7, 19396–19406.
- Kim, H.T.; Hee Ryu, M.; Jung, Y.J.; Lim, S.; Song, H.M.; Park, J.; Hwang, S.Y.; Lee, H.S.; Yeon, Y.J.; Sung, B.H.; et al. Chemo-Biological Upcycling of Poly (ethylene terephthalate) to Multifunctional Coating Materials. ChemSusChem 2021, 14, 4251–4259.
- Kang, M.J.; Kim, H.T.; Lee, M.-W.; Kim, K.-A.; Khang, T.U.; Song, H.M.; Park, S.J.; Joo, J.C.; Cha, H.G. A chemo-microbial hybrid process for the production of 2-pyrone-4, 6-dicarboxylic acid as a promising bioplastic monomer from PET waste. Green Chem. 2020, 22, 3461–3469.
- Werner, A.Z.; Clare, R.; Mand, T.D.; Pardo, I.; Ramirez, K.J.; Haugen, S.J.; Bratti, F.; Dexter, G.N.; Elmore, J.R.; Huenemann, J.D.; et al. Tandem chemical deconstruction and biological upcycling of poly (ethylene terephthalate) to β-ketoadipic acid by Pseudomonas putida KT2440. Metab. Eng. 2021, 67, 250–261.
- Tiso, T.; Narancic, T.; Wei, R.; Pollet, E.; Beagan, N.; Schröder, K.; Honak, A.; Jiang, M.; Kenny, S.T.; Wierckx, N.; et al. Towards bio-upcycling of polyethylene terephthalate. Metab. Eng. 2021, 66, 167–178.
- Sadler, J.C.; Wallace, S. Microbial synthesis of vanillin from waste poly (ethylene terephthalate). Green Chem. 2021, 23, 4665–4672.
- Liu, P.; Zhang, T.; Zheng, Y.; Li, Q.; Su, T.; Qi, Q. Potential one-step strategy for PET degradation and PHB biosynthesis through co-cultivation of two engineered microorganisms. Eng. Microbiol. 2021, 1, 100003.
- Liu, P.; Zheng, Y.; Yuan, Y.; Zhang, T.; Li, Q.; Liang, Q.; Su, T.; Qi, Q. Valorization of Polyethylene Terephthalate to Muconic Acid by Engineering Pseudomonas Putida. Int. J. Mol. Sci. 2022, 23, 10997.
- Mückschel, B.; Simon, O.; Klebensberger, J.; Graf, N.; Rosche, B.; Altenbuchner, J.; Pfannstiel, J.; Huber, A.; Hauer, B. Ethylene glycol metabolism by Pseudomonas putida. Appl. Environ. Microbiol. 2012, 78, 8531–8539.
- Utomo, R.N.C.; Li, W.-J.; Tiso, T.; Eberlein, C.; Doeker, M.; Heipieper, H.J.; Jupke, A.; Wierckx, N.; Blank, L.M. Defined microbial mixed culture for utilization of polyurethane monomers. ACS Sustain. Chem. Eng. 2020, 8, 17466–17474.
- Son, H.F.; Cho, I.J.; Joo, S.; Seo, H.; Sagong, H.-Y.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for highly efficient PET degradation. ACS Catal. 2019, 9, 3519–3526.
- Gautam, R.; Bassi, A.; Yanful, E. A review of biodegradation of synthetic plastic and foams. Appl. Biochem. Biotechnol. 2007, 141, 85–108.
- Magnin, A.; Pollet, E.; Phalip, V.; Avérous, L. Evaluation of biological degradation of polyurethanes. Biotechnol. Adv. 2020, 39, 107457.
- Jin, X.; Dong, J.; Guo, X.; Ding, M.; Bao, R.; Luo, Y. Current advances in polyurethane biodegradation. Polym. Int. 2022, 71, 1384–1392.
- Gamerith, C.; Acero, E.H.; Pellis, A.; Ortner, A.; Vielnascher, R.; Luschnig, D.; Zartl, B.; Haernvall, K.; Zitzenbacher, S.; Strohmeier, G.; et al. Improving enzymatic polyurethane hydrolysis by tuning enzyme sorption. Polym. Degrad. Stab. 2016, 132, 69–77.
- Magnin, A.; Entzmann, L.; Pollet, E.; Avérous, L. Breakthrough in polyurethane bio-recycling: An efficient laccase-mediated system for the degradation of different types of polyurethanes. Waste Manag. 2021, 132, 23–30.
- Branson, Y.; Söltl, S.; Buchmann, C.; Wei, R.; Schaffert, L.; Badenhorst, C.P.; Reisky, L.; Jäger, G.; Bornscheuer, U. Urethanases for the enzymatic hydrolysis of low molecular weight carbamates and the recycling of polyurethanes. Angew. Chem. 2023, e202216220.
- Mi, H.-Y.; Jing, X.; Napiwocki, B.N.; Hagerty, B.S.; Chen, G.; Turng, L.-S. Biocompatible, degradable thermoplastic polyurethane based on polycaprolactone-block-polytetrahydrofuran-block-polycaprolactone copolymers for soft tissue engineering. J. Mater. Chem. B 2017, 5, 4137–4151.
- Jeong, J.; Oh, D.; Goh, M. Synthesis, antibacterial activity, and enzymatic decomposition of bio-polyurethane foams containing propolis. J. Ind. Eng. Chem. 2022, 109, 182–188.
- Liu, J.; He, J.; Xue, R.; Xu, B.; Qian, X.; Xin, F.; Blank, L.M.; Zhou, J.; Wei, R.; Dong, W.; et al. Biodegradation and up-cycling of polyurethanes: Progress, challenges, and prospects. Biotechnol. Adv. 2021, 48, 107730.
- Liu, J.; Zeng, Q.; Lei, H.; Xin, K.; Xu, A.; Wei, R.; Li, D.; Zhou, J.; Dong, W.; Jiang, M.; et al. Biodegradation of polyester polyurethane by Cladosporium sp. P7: Evaluating its degradation capacity and metabolic pathways. J. Hazard. Mater. 2023, 448, 130776.
- Chen, L.; Rong, M.; Yang, L.; Yu, J.; Qu, H.; Meng, Q.; Ni, S.; Xu, Z.; Zhu, X.; Wang, L.; et al. Construction of super-hydrophobic hypercrosslinked porous polymers for selectively removing aromatic diamines from the polyurethane bio-hydrolysate. Chem. Eng. J. 2022, 428, 132509.
- Kotova, I.; Taktarova, Y.V.; Tsavkelova, E.; Egorova, M.; Bubnov, I.; Malakhova, D.; Shirinkina, L.; Sokolova, T.; Bonch-Osmolovskaya, E. Microbial Degradation of Plastics and Approaches to Make it More Efficient. Microbiology 2021, 90, 671–701.
- Palmer, R.J.; Staff, U.B. Polyamides, plastics. In Kirk-Othmer Encyclopedia of Chemical Technology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2005.
- Negoro, S. Biodegradation of nylon oligomers. Appl. Microbiol. Biotechnol. 2000, 54, 461–466.
- Biundo, A.; Subagia, R.; Maurer, M.; Ribitsch, D.; Syrén, P.-O.; Guebitz, G.M. Switched reaction specificity in polyesterases towards amide bond hydrolysis by enzyme engineering. RSC Adv. 2019, 9, 36217–36226.
- Deguchi, T.; Kitaoka, Y.; Kakezawa, M.; Nishida, T. Purification and characterization of a nylon-degrading enzyme. Appl. Environ. Microbiol. 1998, 64, 1366–1371.
- Santo, M.; Weitsman, R.; Sivan, A. The role of the copper-binding enzyme-laccase—In the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeterior. Biodegrad. 2013, 84, 204–210.
- Zhang, J.; Pedersen, N.J.; Eser, B.E.; Guo, Z. Biodegradation of polyethylene and polystyrene: From microbial deterioration to enzyme discovery. Biotechnol. Adv. 2022, 60, 107991.
- Mohanan, N.; Montazer, Z.; Sharma, P.K.; Levin, D.B. Microbial and enzymatic degradation of synthetic plastics. Front. Microbiol. 2020, 11, 580709.
- Eyheraguibel, B.; Traikia, M.; Fontanella, S.; Sancelme, M.; Bonhomme, S.; Fromageot, D.; Lemaire, J.; Lauranson, G.; Lacoste, J.; Delort, A.; et al. Characterization of oxidized oligomers from polyethylene films by mass spectrometry and NMR spectroscopy before and after biodegradation by a Rhodococcus rhodochrous strain. Chemosphere 2017, 184, 366–374.
- Sudhakar, M.; Doble, M.; Murthy, P.S.; Venkatesan, R. Marine microbe-mediated biodegradation of low-and high-density polyethylenes. Int. Biodeterior. Biodegrad. 2008, 61, 203–213.
- Jeon, H.J.; Kim, M.N. Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int. Biodeterior. Biodegrad. 2015, 103, 141–146.
- Yang, S.-S.; Wu, W.-M.; Brandon, A.M.; Fan, H.-Q.; Receveur, J.P.; Li, Y.; Wang, Z.-Y.; Fan, R.; McClellan, R.L.; Gao, S.-H.; et al. Ubiquity of polystyrene digestion and biodegradation within yellow mealworms, larvae of Tenebrio molitor Linnaeus (Coleoptera: Tenebrionidae). Chemosphere 2018, 212, 262–271.
- Harper, C.A. Handbook of Plastics, Elastomers, and Composites; McGraw-Hill: New York, NY, USA, 2002; Volume 4.
- Jain, K.; Bhunia, H.; Sudhakara Reddy, M. Degradation of polypropylene–poly-L-lactide blend by bacteria isolated from compost. Bioremediation J. 2018, 22, 73–90.
- Braun, D. Poly (vinyl chloride) on the way from the 19th century to the 21st century. J. Polym. Sci. Part A Polym. Chem. 2004, 42, 578–586.
- Zhang, Z.; Peng, H.; Yang, D.; Zhang, G.; Zhang, J.; Ju, F. Polyvinyl chloride degradation by a bacterium isolated from the gut of insect larvae. Nat. Commun. 2022, 13, 5360.
- Shah, M.M.; Barr, D.P.; Chung, N.; Aust, S.D. Use of white rot fungi in the degradation of environmental chemicals. Toxicol. Lett. 1992, 64, 493–501.
- Sullivan, K.P.; Werner, A.Z.; Ramirez, K.J.; Ellis, L.D.; Bussard, J.R.; Black, B.A.; Brandner, D.G.; Bratti, F.; Buss, B.L.; Dong, X.; et al. Mixed plastics waste valorization through tandem chemical oxidation and biological funneling. Science 2022, 378, 207–211.
- Guzik, M.W.; Nitkiewicz, T.; Wojnarowska, M.; Sołtysik, M.; Kenny, S.T.; Babu, R.P.; Best, M.; O’Connor, K.E. Robust process for high yield conversion of non-degradable polyethylene to a biodegradable plastic using a chemo-biotechnological approach. Waste Manag. 2021, 135, 60–69.
- Tzouvelekis, L.; Tzelepi, E.; Tassios, P.; Legakis, N. CTX-M-type β-lactamases: An emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 2000, 14, 137–142.
- Wang, W.; Themelis, N.J.; Sun, K.; Bourtsalas, A.C.; Huang, Q.; Zhang, Y.; Wu, Z. Current influence of China’s ban on plastic waste imports. Waste Dispos. Sustain. Energy 2019, 1, 67–78.
- Gruber, F.; Grählert, W.; Wollmann, P.; Kaskel, S. Classification of black plastics waste using fluorescence imaging and machine learning. Recycling 2019, 4, 40.
- Erkinay Ozdemir, M.; Ali, Z.; Subeshan, B.; Asmatulu, E. Applying machine learning approach in recycling. J. Mater. Cycles Waste Manag. 2021, 23, 855–871.
- Zhao, X.; Liu, X.; Feng, K.; An, W.L.; Tian, F.; Du, R.; Xu, S.; Chen, L.; Wu, G.; Wang, Y.Z. Multicycling of Epoxy Thermoset Through a Two-Step Strategy of Alcoholysis and Hydrolysis using a Self-Separating Catalysis System. ChemSusChem 2022, 15, e202101607.
- Platonova, E.; Chechenov, I.; Pavlov, A.; Solodilov, V.; Afanasyev, E.; Shapagin, A.; Polezhaev, A. Thermally Remendable Polyurethane Network Cross-Linked via Reversible Diels-Alder Reaction. Polymers 2021, 13, 1935.
- Zahedifar, P.; Pazdur, L.; Vande Velde, C.M.; Billen, P. Multistage chemical recycling of polyurethanes and dicarbamates: A glycolysis–hydrolysis demonstration. Sustainability 2021, 13, 3583.
- Sivan, A. New perspectives in plastic biodegradation. Curr. Opin. Biotechnol. 2011, 22, 422–426.
- Carmen, S. Microbial capability for the degradation of chemical additives present in petroleum-based plastic products: A review on current status and perspectives. J. Hazard. Mater. 2021, 402, 123534.
- Wei, R.; Tiso, T.; Bertling, J.; O’Connor, K.; Blank, L.M.; Bornscheuer, U.T. Possibilities and limitations of biotechnological plastic degradation and recycling. Nat. Catal. 2020, 3, 867–871.
- Chu, M.; Liu, Y.; Lou, X.; Zhang, Q.; Chen, J. Rational Design of Chemical Catalysis for Plastic Recycling. ACS Catal. 2022, 12, 4659–4679.
- He, J.; Han, L.; Wang, F.; Ma, C.; Cai, Y.; Ma, W.; Xu, E.G.; Xing, B.; Yang, Z. Photocatalytic strategy to mitigate microplastic pollution in aquatic environments: Promising catalysts, efficiencies, mechanisms, and ecological risks. Crit. Rev. Environ. Sci. Technol. 2022, 53, 504–526.
- Jehanno, C.; Alty, J.W.; Roosen, M.; de Meester, S.; Dove, A.P.; Chen, E.Y.-X.; Leibfarth, F.A.; Sardon, H. Critical advances and future opportunities in upcycling commodity polymers. Nature 2022, 603, 803–814.
- Tiso, T.; Winter, B.; Wei, R.; Hee, J.; de Witt, J.; Wierckx, N.; Quicker, P.; Bornscheuer, U.T.; Bardow, A.; Nogales, J.; et al. The metabolic potential of plastics as biotechnological carbon sources—Review and targets for the future. Metab. Eng. 2022, 71, 77–98.
More
Information
Subjects:
Biochemistry & Molecular Biology; Green & Sustainable Science & Technology; Engineering, Environmental
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
2.1K
Revisions:
4 times
(View History)
Update Date:
28 Feb 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




