Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Emily Ann Schiller | -- | 3544 | 2023-02-14 21:48:41 | | | |
| 2 | Catherine Yang | -3 word(s) | 3541 | 2023-02-15 02:23:50 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Schiller, E.A.; Cohen, K.; Lin, X.; El-Khawam, R.; Hanna, N. Extracellular Vesicle-microRNAs as Diagnostic Biomarkers in Preterm Neonates. Encyclopedia. Available online: https://encyclopedia.pub/entry/41232 (accessed on 07 February 2026).
Schiller EA, Cohen K, Lin X, El-Khawam R, Hanna N. Extracellular Vesicle-microRNAs as Diagnostic Biomarkers in Preterm Neonates. Encyclopedia. Available at: https://encyclopedia.pub/entry/41232. Accessed February 07, 2026.
Schiller, Emily A, Koral Cohen, Xinhua Lin, Rania El-Khawam, Nazeeh Hanna. "Extracellular Vesicle-microRNAs as Diagnostic Biomarkers in Preterm Neonates" Encyclopedia, https://encyclopedia.pub/entry/41232 (accessed February 07, 2026).
Schiller, E.A., Cohen, K., Lin, X., El-Khawam, R., & Hanna, N. (2023, February 14). Extracellular Vesicle-microRNAs as Diagnostic Biomarkers in Preterm Neonates. In Encyclopedia. https://encyclopedia.pub/entry/41232
Schiller, Emily A, et al. "Extracellular Vesicle-microRNAs as Diagnostic Biomarkers in Preterm Neonates." Encyclopedia. Web. 14 February, 2023.
Copy Citation
Neonates born prematurely (<37 weeks of gestation) are at a significantly increased risk of developing inflammatory conditions associated with high mortality rates, including necrotizing enterocolitis, bronchopulmonary dysplasia, and hypoxic-ischemic brain damage.
neonate
prematurity
bronchopulmonary dysplasia
necrotizing enterocolitis
1. Definition of an Effective Diagnostic Biomarker
A biomarker is a measurable characteristic that indicates normal physiology, pathological processes, or response to exposure or treatment. The FDA-NIH BEST (Biomarkers, EndpointS, and other Tools) [1] categorizes biomarkers based on their application, including: (1) diagnostic biomarkers to detect the presence of a disease or disease subtype, (2) monitoring biomarkers to assess a parameter over time, (3) predictive biomarkers that identify individuals who are more likely to experience a defined outcome after a specific exposure, (4) prognostic biomarkers that indicate the likelihood of a future clinical event, (5) response biomarkers to show a biological response to exposure, and (6) safety biomarkers that are measured before or after exposure to determine toxicity. Understanding these definitions is imperative for identifying clinically useful biomarkers.
Ideal biomarkers should have the following characteristics [1][2][3]: (1) present in peripheral tissues or fluids that are suitable for sample collection from the target patient population and are involved in the pathophysiological process of the disease; (2) present at a sufficiently high concentration to be detected within a reasonable, defined amount of sample; (3) measurable quickly and affordably with robust analytic performance across various clinical settings; and (4) highly sensitive and specific for the disease in the target population and able to differentiate between diseases that might have similar clinical presentations.
In neonates, there is an urgent need for biomarker discovery to inform and enable early decision-making and personalized treatment plans. Previous approaches aimed at the identification of such biomarkers in neonates have been largely limited by several factors, including: (1) attempting to predict a multi-factorial disease that has diverse pathophysiology by focusing on biomarkers involved in only one particular pathway; (2) the difficulty of identifying a noninvasive sampling site that can accurately mirror biological processes occurring in a specific organ; (3) trying to identify biomarkers that distinguish disease processes that are too advanced in the disease course, limiting effective intervention early in the disease process; (4) using non-sensitive detection techniques, or the use of an intricate assay used only in a research lab that cannot be transferred to a clinically applicable assay; and (5) lack of validation of biomarker expression in larger patient cohorts [4][5][6]. These issues are compounded by the delicate clinical status and small blood volume of neonates, which preclude frequent blood draws for biomarker assessment. EVs obtained from different non-invasive biofluids can be exploited as accurate biomarkers representative of distinct pathological pathways identified early in the disease process.
2. EVs as Effective Diagnostic Biomarkers
EVs, which are released by all cells and are ubiquitous in all bodily fluids, are recognized as highly efficient and biologically significant intercellular communication systems [7][8]. EVs are membrane-bound vesicles secreted by cells to mediate cell signaling and deliver cellular contents to target cells. The targeting and uptake of EVs can be specific or non-specific, depending on their protein and lipid composition [9]. There are two major subtypes of EVs: (1) exosomes (50–150 nm in diameter), which form through the fusion of intraluminal vesicle-containing multivesicular bodies with the plasma membrane [10][11] and (2) microvesicles (50–500 nm in diameter), which form through outward blebbing of the plasma membrane [10][11]. Because exosomes originate from the endocytic compartment, their molecular content mainly reflects that of the parental cell [10]; they serve as surrogates of their cells of origin and are recognized as “liquid biopsies” [12][13]. EVs contain various metabolites, nucleic acids, and proteins that alter cell signaling, protein regulation, and gene expression in target cells [14][15].
EVs are found in various biological fluids, including peripheral blood, umbilical cord blood, saliva, urine, tears, tracheal fluid, and breast milk [15][16][17][18][19]. In addition, EVs can be purified and enriched from these biological fluids to detect EVs and miRNAs that were previously too small in quantity to be identified. This provides researchers with an optimal opportunity to study the EV content associated with various disease processes. EV-miRNAs serve as candidate biomarkers for many diseases, with most studies focusing on their role in cancer diagnosis [13][20]. Increasing reports show that the sorting of miRNAs is an active process. As such, EV-miRNAs reflect the status of the cells from which they are secreted, and a diseased state can be revealed by sampling biological fluids instead of performing a biopsy on pathologic tissues [21][22][23][24][25][26]. Furthermore, EVs derived from pathological tissues may express different surface markers, enabling the specific isolation of such EVs [21].
Methods of EV Characterization and miRNA Extraction
To develop EV biomarkers, characterization of EVs from target biofluid and quantitative and qualitative analysis of the EV cargos are essential. The concentration, size, and surface zeta potential can be assayed by nanoparticle tracking analysis (NTA), such as Nanosight and Zetaview [27][28]. To further investigate the protein cargos, EVs can be analyzed by single particle interferometric reflectance imaging sensor (SP-IRIS) using the Exo-View platform [28][29]. To characterize EV morphology, electron microscopy (EM) is commonly used. EM analysis can observe the lipid bilayer and differentiate EVs from dense particles such as lipoproteins [30].
More advanced analytical methods have been developed to study morphology in more detail. Hardij et al. introduced atomic force microscopy as an alternative method for visualizing EVs [31]. Using this technique, it is possible to visualize a single EV and the specific surface antigens. Raman microspectroscopy has also been described as an alternative for label-free visualization of EVs [32]. Using a detection platform that combines a microfluidic device and surface-enhanced Raman spectroscopy (SERS), Wang et al. were able to profile four protein biomarkers in serum EVs [32]. Recently, holotomography imaging has been introduced to gain new insights into EV characterization with an optical, contact-free, label-free examination [33]. Conventional protein analysis techniques such as western blots and ELISA can be used to determine the EV fraction’s protein cargo level.
To quantify miRNAs in biological fluids and EVs, total RNA or RNA with small RNA enrichment extraction is performed with RNA extraction kits, such as column-based extraction [34], chloroform–phenol-based extraction [35], magnetic bead extraction [35], then microarray [36], Northern blotting [37], and quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis [38]. Among these methods, qRT-PCR is widely preferred over other detection methods because of its high sensitivity and specificity for detecting low levels of circulating miRNAs in plasma and serum. In the qRT-PCR method, cDNA from specific miRNA is reverse transcribed using specific stem-loop RT primers [39][40] or with the poly-A adopter approach [41][42], followed by PCR with specific PCR primers. Since qRT-PCR is a standard technique already employed in research and central clinical laboratories, it is usually the method of choice in the initial discovery and assay development stages. However, the requirement of reverse transcription and indirect measurement renders the qRT-PCR methods limited in robustness and accuracy. In addition, it is extremely complicated, time-consuming, and laborious; as such, it is unsuitable for clinical practice, particularly in a point-of-care setting. Optical fluorescence-based biosensors that detect the hybridization between the miRNAs and their respective complementary mRNA probes are highly sensitive using fluorescence spectroscopy [43][44]. The label-free detection of biomolecules has been a long-standing goal in developing optical biosensors [45][46][47]. The working principle of the biosensor is measuring the change in the intrinsic physical parameter of the biosensor caused by the binding of miRNA molecules. Therefore, the biosensor methods can assay the target miRNA in its natural state, unmodified. This results in a cost-effective, more reliable, easy, and faster real-time biorecognition interaction detection. Another advantage of the biosensor platform is the ultra-small detection volume requirement and extremely low detection limit (down to the attomole level in some cases). A more detailed discussion of the biosensor platform in miRNA detection is beyond the scope of this paper; readers are recommended to consult reviews by Zhang et al. [48], Dave et al. [49], Cacheux et al. [50], and Lai and Slaughter [51].
3. Next Steps in EV-miRNAs Biomarker Development in Premature Infants
EV-miRNAs have the potential to serve as diagnostic biomarkers for conditions affecting premature neonates, such as NEC, BPD, and HIBD. However, research in this area is preliminary, and many challenges must be addressed before this work can be translated into clinical practice. The major challenges are discussed below and illustrated in Figure 1.
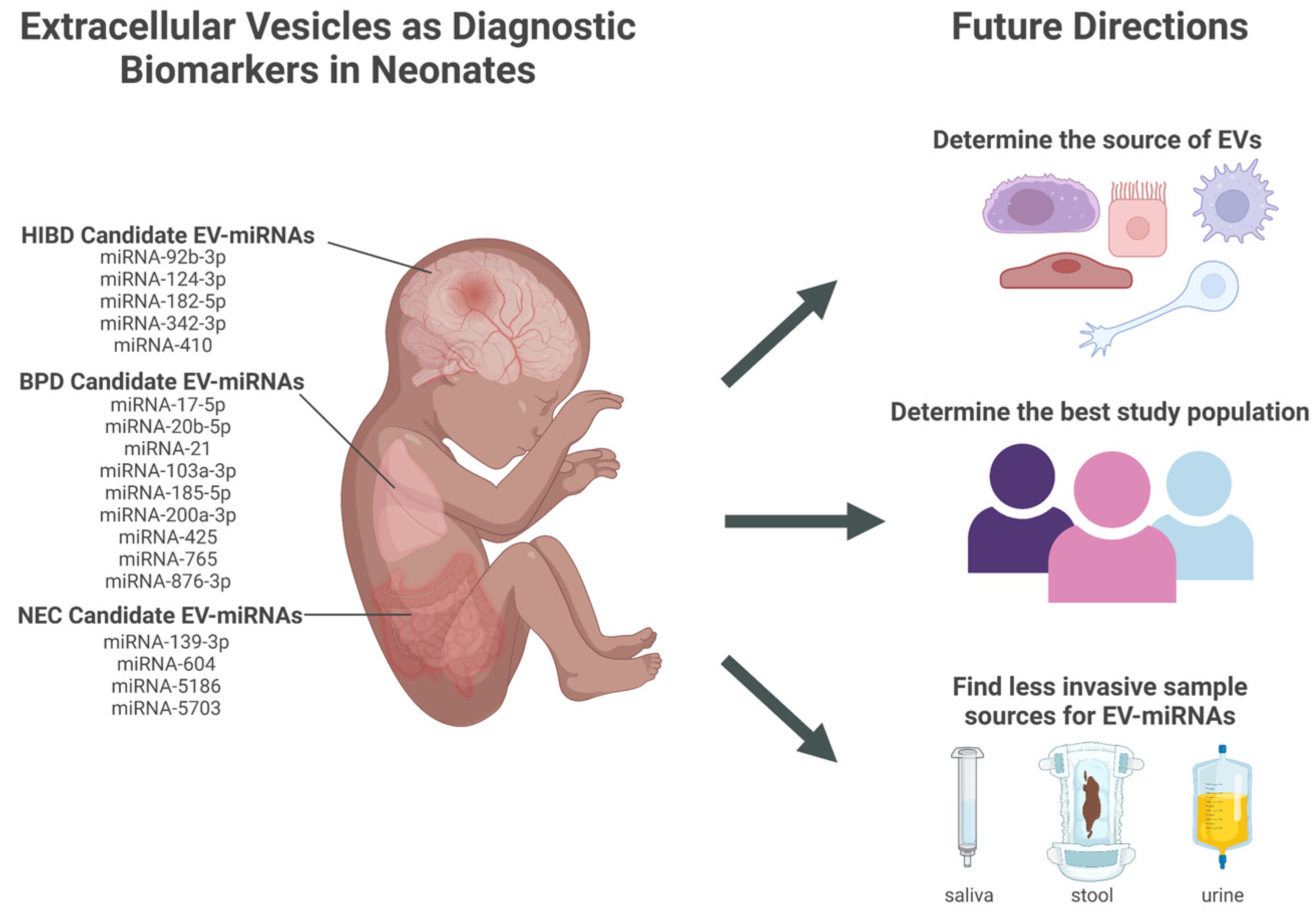
Figure 1. Candidate extracellular vesicles (EVs) and their miRNAs content as diagnostic biomarkers for neonatal conditions and future directions that address challenges in translation to clinical practice. EVs (extracellular vesicles); miRNA (micro-RNA); HIBD (hypoxic-ischemic brain damage); BPD (bronchopulmonary dysplasia); NEC (necrotizing enterocolitis). Created in BioRender.com.
3.1. Determine the Best Sample Source for Discovering Diagnostic Biomarkers from Neonatal EVs
The source of the biomarkers is crucial when considering how the research presented here will translate into clinical medicine. EVs can be isolated from many sources, such as blood (serum or plasma), urine, saliva, and feces. There are various methods of isolating EVs, which include ultracentrifugation, precipitation, and size exclusion chromatography (SEC) [52][53]. Long processing time, lack of specificity and sensitivity, and high cost are among some of the limitations of these methods [54]. While EVs can serve as diagnostic biomarkers, there are some limitations that may hinder their use, which may be due to the way they are isolated.
Due to their heterogeneous nature, EVs can be very difficult to quantify [55]. Quantification methods include NTA, dynamic light scattering, and tunable resistive pulse sensing [56]. However, these methods have limited use due to their inability to distinguish between lipoproteins and particles of protein aggregates from EVs [57]. Plasma, which has an abundant source of lipoproteins and aggregates of protein, is a biofluid that requires different techniques to quantify and isolate EVs [58]. One feature of EVs that may aid in their quantification and isolation is the presence of transmembrane proteins. These transmembrane proteins may act as EV markers, making them useful during isolation and quantification.
One studied method of isolating EVs is an insulator-based dielectrophoretic device that is capable of isolating EVs from small sample volumes with a short processing time [54]. Another studied method of isolating EVs is advanced mass spectrometry (MS), which is able to distinguish the protein content of EVs under various physiologic and pathologic conditions [59]. Since EVs can reflect the content of their cells of origin, this serves to be beneficial when deciding which biofluid to analyze in specific neonatal disease processes. One of the major hindrances to characterizing EVs in different biofluids, such as urine and blood, is the presence of a higher magnitude of proteins when compared to EVs [60]. As a result, additional isolation methods are used prior to MS to better extract and characterize EVs. Some of these isolation methods focus on the physical property, such as size and density, as well as EVs’ chemical properties, to better isolate EVs [60].
The characterization of EVs from different biofluids can be affected by many factors, including improper storage and processing conditions. One of the major biofluids used to study appropriate storage, collection, and processing conditions is blood, or, more specifically, plasma. The use of anticoagulants when using blood as the biofluid for EV analysis is controversial. Heparin-based anticoagulants are discouraged. Heparin is associated with false-negative PCR readings since heparin competes with enzymes needed for binding to nucleic acid and can bind to EVs as well as block their uptake [52]. Another factor that can play a role in the optimal isolation of EVs is the fasting state of the patient. Some will analyze the blood samples fairly quickly within one hour of collection, while others will collect blood samples after a 12-hour fasting period [52][60]. This is believed to be required for accurate EV acquirement. The storage of biofluid samples is also controversial. Many have stored samples at 4 °C for up to 5 days without any effect on the number of EVs isolated [61]. However, for long-term storage, samples should be frozen at or below −80 °C and repeated freeze-thaw cycles must be avoided to maintain sample integrity [52].
When choosing which biofluid to analyze in neonates, the ease of collection also comes into play. In the neonatal population, due to their low blood volumes and their susceptibility to becoming anemic with even the smallest of blood draws, blood may not be the ideal biofluid to use when trying to identify biomarkers of diseases. As a result, less invasive biofluid samples, such as urine, saliva, or feces, should become more in favor when analyzing and isolating EVs for potential use as a biomarker. Research on EV isolation in feces is limited. A recent study aimed to address this gap in knowledge through the comparison of EV-isolation techniques in healthy adults [61]. In this study, Tris-EDTA-based preservative buffer was added to stool samples, and the samples were centrifuged and vortexed prior to storage at −80 °C. For EV isolation, ultracentrifugation, precipitation, SEC, and ultrafiltration were compared. It was observed that SEC was the method of choice when considering recovery, reproducibility, and purity. Regarding the neonatal population, there are potential miRNA biomarkers for NEC in neonatal fecal samples [62], and EVs have recently been isolated from the first-pass meconium [63]. However, whether EVs are specifically present in preterm neonatal feces and whether EV-specific miRNAs can serve as biomarkers for NEC or other neonatal diseases remain unknown.
Furthermore, EVs have been characterized in neonatal urine [64]. Numerous studies have been published in adults detailing collection and storage protocols to maximize the stability and recovery of urinary EVs, though there is not one standard protocol that has been established. A recent review article analyzing methods for urine EV isolation concluded that once the urine is collected, it should be stored between 0–4 °C and processed within 8 h [65]. During processing, samples undergo centrifugation to remove cells, cellular debris, and urinary protein uromodulin [65]. Urine samples are then stored at −80 °C, a temperature at which EV-miRNAs are stable even after long-term storage [65][66]. There is conflicting evidence on whether protease inhibitors should be added to samples prior to freezing to prevent urinary EV degradation [66]. However, the use of protease inhibitors would substantially increase the cost of urine biomarker discovery [65]. In Galley et al., urine was collected from preterm neonates by placing cotton balls in their diapers [64] and the urine samples were frozen at −80 °C without processing or the addition of a protease inhibitor. Samples were thawed prior to EV isolation. Urine sample collection in the neonatal population can be challenging as neonates cannot time their voids to easily coordinate a clean-catch sample. Additionally, placing catheters for sterile urine collection introduces the risk of urinary tract infections, which can be particularly dangerous in the preterm population. While Galley et al. were able to isolate and characterize preterm neonatal urine EVs, it is important for future work to develop a standard protocol for urinary EV collection and storage, particularly in the neonatal population.
We were unable to identify studies in which EVs were obtained from neonatal saliva, although their presence in adult saliva [67][68] suggests that EVs are likely to be present in neonatal saliva. The small amount of saliva produced and the inability of neonates to voluntarily provide a sample, pose a potential challenge in sample collection for biomarker discovery. However, a simple bedside suction technique yields between 10 and 50 μL and can be done in extremely premature neonates [6]. Once saliva is collected, commercially available stabilizing solutions can be utilized in this population [69]. Another potential challenge is the effects of hydration status in saliva production, limiting the ability to normalize samples based on volume. To address this, one research group determined that GAPDH, YWHAZ, and HPRT1 are the optimal reference genes for RT-qPCR normalization in neonates as they maintain their stability across various gestational and post-menstrual ages [6][70]. Overall, previous work in neonatal saliva collection and preparation suggests that it can be utilized for EV-miRNAs biomarker discovery.
Many techniques are used to store, process, isolate, and characterize EVs from various biofluids to obtain an accurate yield that will best be used as a clinical biomarker in various disease processes in the neonatal population. Moreover, the disease process may drive which biofluid would be collected for EV isolation and analysis. It is imperative to understand the cellular origin of EVs present in different tissue samples and how enriched pathological tissue-derived EVs in a sample affect their performance as a diagnostic biomarker. A recent study utilizing adult plasma demonstrated that 99.8% of plasma EVs originated from hematopoietic cells and that the remaining 0.2% originated from other tissues [71]. Interestingly, the fraction of EVs derived from liver cells increased in the plasma of patients with hepatocellular carcinoma, indicating that the cell-origin profile of EVs may reflect disease states [71]. Therefore, the cell-origin profile of EVs in a particular sample, by its very nature, has the potential to serve as a diagnostic biomarker. For example, to determine an accurate biomarker for NEC, one may choose to collect and analyze stool, and to determine an accurate biomarker for BPD, one may choose to collect saliva or tracheal aspirates to better obtain and isolate EVs that are more specific to lung pathology. However, to the knowledge, no study has described the cell-origin profile of EVs in neonatal samples.
Additionally, while hUC-EVs may serve as a diagnostic biomarker in specific neonatal diseases, hUC has the major drawback of only providing data from a single time point in a premature neonate’s hospital course. Biomarkers obtained from a single time point with no option to repeat as needed have limited value as diagnostic and prognostic tools in progressive diseases or for monitoring response to treatment.
3.2. Validation of EV-miRNAs as Reliable Diagnostic Biomarkers in the Premature Population
As evidenced by the work summarized here, evidence for the use of EV-miRNAs as diagnostic biomarkers is preliminary, with the goal of discovering candidate biomarkers rather than validating the clinical use of such biomarkers. For example, the studies described here have limited sample sizes, with samples obtained from a single center. While most studies reported -p-values between experimental and control group EV-miRNAs expression, only two studies reported an area under the curve. Therefore, multi-center clinical studies with larger sample sizes that assess metrics such as sensitivity, specificity, area under the curve, positive and negative protective values, etc., are required to further understand whether the miRNAs discussed here are reliable biomarkers.
As work in preterm EV-miRNAs advances and robust clinical studies are designed, there will be several variables to consider. Determining the right patient population for biomarker discovery and validation will be critical in the clinical application of future work. As evidenced by the work presented here, there is no standardization in what gestational age to include in the preterm population. For example, while some studies for BPD included all neonates <32 weeks gestation, others only included neonates <28 weeks gestation. Meanwhile, prematurity is clinically defined as <37 weeks gestation. Since the incidence of NEC and BPD increases as gestational age decreases, will narrowing the gestational age range to those who are extremely premature lead to higher diagnostic accuracy? Additionally, it is unclear how controls will be defined. While some studies here utilized premature neonates without the condition being studied, others used full-term controls . Since NEC and BPD are extremely rare pathologies in the full-term neonate, future work should focus on utilizing only preterm neonates as controls.
Furthermore, there is much work to be done exploring EV-miRNAs in preterm HIBD. Most studies not only focus on the full-term population but also do not specifically isolate miRNAs from EVs. While other studies have identified the presence of such HIBD miRNAs within EVs, future studies must be conducted to compare EV-miRNAs between preterm neonates with and without HIBD. This will not only further explore the diagnostic potential of the candidate miRNAs but also identify other candidates EV-miRNAs for validation studies.
3.3. Determine the Feasibility of Implementing EV Diagnostic Testing: Testing Population and Role of EV Biomarkers in Clinical Decision Making
Unnecessary laboratory studies increase the risk of negative outcomes and increase the cost of hospitalization, especially when test results lead providers to initiate unnecessary interventions [72][73]. Therefore, future biomarker discovery should focus on a subpopulation of high-risk premature neonates to implement EV-based diagnostic testing. Whether providers should perform EV analysis for NEC or BPD in all premature neonates or only in those who are at high risk of developing such pathology needs to be considered in future studies.
Because the pathogenesis of some diseases in neonates, such as NEC or BPD, is multi-factorial, specific biomarkers may prove to be useful in following disease progression, as well as in direct evaluations and therapeutic options toward a particular pathway of the disease. For example, BPD has been proposed to be the end result of a cascade of events, which may begin with oxygen toxicity, ventilator volume trauma, intrauterine or post-natal infection, and inflammation [74]. Each of these mechanisms contributes to BPD development. Therefore, it is unlikely that the mediators involved in the cascade are identical regardless of the underlying etiology. Using multiple biomarkers from different and distinct biological pathways may differentiate the inciting event and allow pathway-specific therapy directed at the underlying cause of neonatal diseases [75].
The use of EVs and their contents to diagnose conditions in premature neonates may revolutionize laboratory workups and medical interventions in the NICU. However, studies have not specifically analyzed whether the presence of EVs or their contents can aid in clinical decision-making. Researchers are currently designing tools for integration into electronic medical systems that predict NICU length of stay [76], development of BPD [77], NEC vs. spontaneous intestinal perforation [78], sepsis risk [79], and discharge with nasogastric/gastric tube placement [80] in premature neonates. Therefore, future work should analyze whether EVs can provide data points in such tools, contributing to how NICU providers manage care.
References
- FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and Other Tools) Resource. Available online: https://www.ncbi.nlm.nih.gov/books/NBK326791/ (accessed on 16 September 2022).
- Byrnes, S.A.; Weigl, B.H. Selecting Analytical Biomarkers for Diagnostic Applications: A First Principles Approach. Expert Rev. Mol. Diagn. 2017, 18, 19–26.
- Califf, R.M. Biomarker Definitions and Their Applications. Exp. Biol. Med. 2018, 243, 213–221.
- Murphy, C.A.; O’Reilly, D.P.; Neary, E.; EL-Khuffash, A.; NíAinle, F.; McCallion, N.; Maguire, P.B. A Review of the Role of Extracellular Vesicles in Neonatal Physiology and Pathology. Pediatr. Res. 2021, 90, 289–299.
- Agakidou, E.; Agakidis, C.; Gika, H.; Sarafidis, K. Emerging Biomarkers for Prediction and Early Diagnosis of Necrotizing Enterocolitis in the Era of Metabolomics and Proteomics. Front. Pediatr. 2020, 8, 602255.
- Yen, E.; Kaneko-Tarui, T.; Maron, J.L. Technical Considerations and Protocol Optimization for Neonatal Salivary Biomarker Discovery and Analysis. Front. Pediatr. 2021, 8, 618553.
- Whiteside, T.L. Extracellular Vesicles Isolation and Their Biomarker Potential: Are We Ready for Testing? Ann. Transl. Med. 2017, 5, 54.
- Boyiadzis, M.; Whiteside, T.L. Information Transfer by Exosomes: A New Frontier in Hematologic Malignancies. Blood Rev. 2015, 29, 281–290.
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606.
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228.
- Raposo, G.; Stahl, P.D. Extracellular Vesicles: A New Communication Paradigm? Nat. Rev. Mol. Cell Biol. 2019, 20, 509–510.
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol. Neurobiol. 2016, 36, 301–312.
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid Biopsy of Cancer: A Multimodal Diagnostic Tool in Clinical Oncology. Ther. Adv. Med. Oncol. 2018, 10, 1758835918794630.
- Gurunathan, S.; Kang, M.-H.; Kim, J.-H. A Comprehensive Review on Factors Influences Biogenesis, Functions, Therapeutic and Clinical Implications of Exosomes. Int. J. Nanomed. 2021, 16, 1281–1312.
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367.
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.-J.; Lahesmaa, R.; Norman, M.; Neve, E.P.A.; Scheynius, A.; Gabrielsson, S. Exosomes with Immune Modulatory Features Are Present in Human Breast Milk. J. Immunol. 2007, 179, 1969–1978.
- Liu, J.; Sun, W.; Liu, C.; Na, Q. Umbilical Cord Blood-Derived Exosomes in Maternal–Fetal Disease: A Review. Reprod. Sci. 2022, 30, 1–8.
- Michael, A.; Bajracharya, S.; Yuen, P.; Zhou, H.; Star, R.; Illei, G.; Alevizos, I. Exosomes from Human Saliva as a Source of MicroRNA Biomarkers. Oral. Dis. 2010, 16, 34–38.
- Keller, S.; Rupp, C.; Stoeck, A.; Runz, S.; Fogel, M.; Lugert, S.; Hager, H.-D.; Abdel-Bakky, M.S.; Gutwein, P.; Altevogt, P. CD24 Is a Marker of Exosomes Secreted into Urine and Amniotic Fluid. Kidney Int. 2007, 72, 1095–1102.
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. New Engl. J. Med. 2018, 379, 2179–2181.
- Ciferri, M.C.; Quarto, R.; Tasso, R. Extracellular Vesicles as Biomarkers and Therapeutic Tools: From Pre-Clinical to Clinical Applications. Biology 2021, 10, 359.
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232.
- Linxweiler, J.; Junker, K. Extracellular Vesicles in Urological Malignancies: An Update. Nat. Rev. Urol. 2020, 17, 11–27.
- Temoche-Diaz, M.M.; Shurtleff, M.J.; Nottingham, R.M.; Yao, J.; Fadadu, R.P.; Lambowitz, A.M.; Schekman, R. Distinct Mechanisms of MicroRNA Sorting into Cancer Cell-Derived Extracellular Vesicle Subtypes. Elife 2019, 8, e47544.
- Lee, H.; Li, C.; Zhang, Y.; Zhang, D.; Otterbein, L.E.; Jin, Y. Caveolin-1 Selectively Regulates MicroRNA Sorting into Microvesicles after Noxious Stimuli. J. Exp. Med. 2019, 216, 2202–2220.
- Martins-Marques, T.; Costa, M.C.; Catarino, S.; Simoes, I.; Aasen, T.; Enguita, F.J.; Girao, H. Cx43-mediated Sorting of MiRNAs into Extracellular Vesicles. Embo Rep. 2022, 23, e54312.
- Bachurski, D.; Schuldner, M.; Nguyen, P.-H.; Malz, A.; Reiners, K.S.; Grenzi, P.C.; Babatz, F.; Schauss, A.C.; Hansen, H.P.; Hallek, M.; et al. Extracellular Vesicle Measurements with Nanoparticle Tracking Analysis—An Accuracy and Repeatability Comparison between NanoSight NS300 and ZetaView. J. Extracell Vesicles 2019, 8, 1596016.
- Tiozzo, C.; Bustoros, M.; Lin, X.; Mejia, C.M.D.; Gurzenda, E.; Chavez, M.; Hanna, I.; Aguiari, P.; Perin, L.; Hanna, N. Placental Extracellular Vesicles–Associated MicroRNA-519c Mediates Endotoxin Adaptation in Pregnancy. Am. J. Obstet. Gynecol. 2021, 225, 681.e1–681.e20.
- Daaboul, G.G.; Gagni, P.; Benussi, L.; Bettotti, P.; Ciani, M.; Cretich, M.; Freedman, D.S.; Ghidoni, R.; Ozkumur, A.Y.; Piotto, C.; et al. Digital Detection of Exosomes by Interferometric Imaging. Sci. Rep. 2016, 6, 37246.
- Berger, A.; Araújo-Filho, I.; Piffoux, M.; Nicolás-Boluda, A.; Grangier, A.; Boucenna, I.; Real, C.C.; Marques, F.L.N.; Faria, D.d.P.; do Rego, A.C.M.; et al. Local Administration of Stem Cell-Derived Extracellular Vesicles in a Thermoresponsive Hydrogel Promotes a pro-Healing Effect in a Rat Model of Colo-Cutaneous Post-Surgical Fistula. Nanoscale 2020, 13, 218–232.
- Hardij, J.; Cecchet, F.; Berquand, A.; Gheldof, D.; Chatelain, C.; Mullier, F.; Chatelain, B.; Dogné, J. Characterisation of Tissue Factor-bearing Extracellular Vesicles with AFM: Comparison of Air-tapping-mode AFM and Liquid Peak Force AFM. J. Extracell Vesicles 2013, 2, 21045.
- Wang, J.; Kao, Y.; Zhou, Q.; Wuethrich, A.; Stark, M.S.; Schaider, H.; Soyer, H.P.; Lin, L.L.; Trau, M. An Integrated Microfluidic-SERS Platform Enables Sensitive Phenotyping of Serum Extracellular Vesicles in Early Stage Melanomas. Adv. Funct. Mater. 2022, 32, 2010296.
- Zadka, Ł.; Buzalewicz, I.; Ulatowska-Jarża, A.; Rusak, A.; Kochel, M.; Ceremuga, I.; Dzięgiel, P. Label-Free Quantitative Phase Imaging Reveals Spatial Heterogeneity of Extracellular Vesicles in Select Colon Disorders. Am. J. Pathol. 2021, 191, 2147–2171.
- Guo, Y.; Vickers, K.; Xiong, Y.; Zhao, S.; Sheng, Q.; Zhang, P.; Zhou, W.; Flynn, C.R. Comprehensive Evaluation of Extracellular Small RNA Isolation Methods from Serum in High Throughput Sequencing. Bmc Genom. 2017, 18, 50.
- Kroh, E.M.; Parkin, R.K.; Mitchell, P.S.; Tewari, M. Analysis of Circulating MicroRNA Biomarkers in Plasma and Serum Using Quantitative Reverse Transcription-PCR (QRT-PCR). Methods 2010, 50, 298–301.
- Zhu, W.; Su, X.; Gao, X.; Dai, Z.; Zou, X. A Label-Free and PCR-Free Electrochemical Assay for Multiplexed MicroRNA Profiles by Ligase Chain Reaction Coupling with Quantum Dots Barcodes. Biosens. Bioelectron. 2014, 53, 414–419.
- Pall, G.S.; Hamilton, A.J. Improved Northern Blot Method for Enhanced Detection of Small RNA. Nat. Protoc. 2008, 3, 1077–1084.
- Zöllner, H.; Hahn, S.A.; Maghnouj, A. Quantitative RT-PCR Specific for Precursor and Mature MiRNAs. Methods Mol. Biol. Clifton N. J. 2013, 1095, 121–134.
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-Time Quantification of MicroRNAs by Stem–Loop RT–PCR. Nucleic Acids Res. 2005, 33, e179.
- Kappel, A.; Keller, A. MiRNA Assays in the Clinical Laboratory: Workflow, Detection Technologies and Automation Aspects. Clin. Chem. Lab. Med. Cclm 2017, 55, 636–647.
- Shi, R.; Chiang, V.L. Facile Means for Quantifying MicroRNA Expression by Real-Time PCR. Biotechniques 2005, 39, 519–525.
- Fiedler, S.D.; Carletti, M.Z.; Christenson, L.K. Quantitative RT-PCR Methods for Mature MicroRNA Expression Analysis. Methods Mol. Biology Clifton N. J. 2010, 630, 49–64.
- Carrascosa, L.G.; Huertas, C.S.; Lechuga, L.M. Prospects of Optical Biosensors for Emerging Label-Free RNA Analysis. Trac. Trends Anal. Chem. 2016, 80, 177–189.
- Giuliano, K.A.; Taylor, D.L. Fluorescent-Protein Biosensors: New Tools for Drug Discovery. Trends Biotechnol. 1998, 16, 135–140.
- Sang, S.; Wang, Y.; Feng, Q.; Wei, Y.; Ji, J.; Zhang, W. Progress of New Label-Free Techniques for Biosensors: A Review. Crit. Rev. Biotechnol. 2016, 36, 465–481.
- Cooper, M.A. Optical Biosensors in Drug Discovery. Nat. Rev. Drug Discov. 2002, 1, 515–528.
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive Optical Biosensors for Unlabeled Targets: A Review. Anal. Chim. Acta 2008, 620, 8–26.
- Zhang, L.; Su, W.; Liu, S.; Huang, C.; Ghalandari, B.; Divsalar, A.; Ding, X. Recent Progresses in Electrochemical DNA Biosensors for MicroRNA Detection. Phenomics 2022, 2, 18–32.
- Dave, V.P.; Ngo, T.A.; Pernestig, A.-K.; Tilevik, D.; Kant, K.; Nguyen, T.; Wolff, A.; Bang, D.D. MicroRNA Amplification and Detection Technologies: Opportunities and Challenges for Point of Care Diagnostics. Lab. Investig. 2019, 99, 452–469.
- Cacheux, J.; Bancaud, A.; Leichlé, T.; Cordelier, P. Technological Challenges and Future Issues for the Detection of Circulating MicroRNAs in Patients With Cancer. Front. Chem. 2019, 7, 815.
- Lai, M.; Slaughter, G. Label-Free MicroRNA Optical Biosensors. Nanomaterials 2019, 9, 1573.
- Coumans, F.A.W.; Brisson, A.R.; Buzas, E.I.; Dignat-George, F.; Drees, E.E.E.; El-Andaloussi, S.; Emanueli, C.; Gasecka, A.; Hendrix, A.; Hill, A.F.; et al. Methodological Guidelines to Study Extracellular Vesicles. Circ. Res. 2017, 120, 1632–1648.
- Royo, F.; Théry, C.; Falcón-Pérez, J.M.; Nieuwland, R.; Witwer, K.W. Methods for Separation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey Performed by the ISEV Rigor and Standardization Subcommittee. Cells 2020, 9, 1955.
- Shi, L.; Kuhnell, D.; Borra, V.J.; Langevin, S.M.; Nakamura, T.; Esfandiari, L. Rapid and Label-Free Isolation of Small Extracellular Vesicles from Biofluids Utilizing a Novel Insulator Based Dielectrophoretic Device. Lab. Chip. 2019, 19, 3726–3734.
- Tkach, M.; Kowal, J.; Théry, C. Why the Need and How to Approach the Functional Diversity of Extracellular Vesicles. Philos. Trans. R. Soc B Biological Sci. 2018, 373, 20160479.
- Hartjes, T.A.; Mytnyk, S.; Jenster, G.W.; van Steijn, V.; Royen, M.E. van Extracellular Vesicle Quantification and Characterization: Common Methods and Emerging Approaches. Bioengineering 2019, 6, 7.
- Simonsen, J.B. What Are We Looking At? Extracellular Vesicles, Lipoproteins, or Both? Circ. Res. 2017, 121, 920–922.
- Osteikoetxea, X.; Sódar, B.; Németh, A.; Szabó-Taylor, K.; Pálóczi, K.; Vukman, K.V.; Tamási, V.; Balogh, A.; Kittel, Á.; Pállinger, É.; et al. Differential Detergent Sensitivity of Extracellular Vesicle Subpopulations. Org. Biomol. Chem. 2015, 13, 9775–9782.
- Li, J.; He, X.; Deng, Y.; Yang, C. An Update on Isolation Methods for Proteomic Studies of Extracellular Vesicles in Biofluids. Molecules 2019, 24, 3516.
- SZATANEK, R.; BARAN, J.; SIEDLAR, M.; BAJ-KRZYWORZEKA, M. Isolation of Extracellular Vesicles: Determining the Correct Approach (Review). Int. J. Mol. Med. 2015, 36, 11–17.
- Northrop-Albrecht, E.J.; Taylor, W.R.; Huang, B.Q.; Kisiel, J.B.; Lucien, F. Assessment of Extracellular Vesicle Isolation Methods from Human Stool Supernatant. J. Extracell Vesicles 2022, 11, e12208.
- Ng, P.C.; Chan, K.Y.Y.; Lam, H.S.; Wong, R.P.O.; Ma, T.P.Y.; Sit, T.; Leung, K.T.; Chan, L.C.N.; Pang, Y.L.I.; Cheung, H.M.; et al. A Prospective Cohort Study of Fecal MiR-223 and MiR-451a as Noninvasive and Specific Biomarkers for Diagnosis of Necrotizing Enterocolitis in Preterm Infants. Neonatology 2021, 117, 555–561.
- Turunen, J.; Tejesvi, M.V.; Suokas, M.; Virtanen, N.; Paalanne, N.; Kaisanlahti, A.; Reunanen, J.; Tapiainen, T. Bacterial Extracellular Vesicles in the Microbiome of First-Pass Meconium in Newborn Infants. Pediatr. Res. 2022, 1–10.
- Galley, J.D.; Mar, P.; Wang, Y.; Han, R.; Rajab, A.; Besner, G.E. Urine-Derived Extracellular Vesicle MiRNAs as Possible Biomarkers for and Mediators of Necrotizing Enterocolitis: A Proof of Concept Study. J. Pediatr. Surg. 2021, 56, 1966–1975.
- Erdbrügger, U.; Blijdorp, C.J.; Bijnsdorp, I.V.; Borràs, F.E.; Burger, D.; Bussolati, B.; Byrd, J.B.; Clayton, A.; Dear, J.W.; Falcón-Pérez, J.M.; et al. Urinary Extracellular Vesicles: A Position Paper by the Urine Task Force of the International Society for Extracellular Vesicles. J. Extracell Vesicles 2021, 10, e12093.
- Barreiro, K.; Dwivedi, O.P.; Valkonen, S.; Groop, P.; Tuomi, T.; Holthofer, H.; Rannikko, A.; Yliperttula, M.; Siljander, P.; Laitinen, S.; et al. Urinary Extracellular Vesicles: Assessment of Pre-analytical Variables and Development of a Quality Control with Focus on Transcriptomic Biomarker Research. J. Extracell Vesicles 2021, 10, e12158.
- Cheshmi, B.; Cheshomi, H. Salivary Exosomes: Properties, Medical Applications, and Isolation Methods. Mol. Biol. Rep. 2020, 47, 6295–6307.
- Han, Y.; Jia, L.; Zheng, Y.; Li, W. Salivary Exosomes: Emerging Roles in Systemic Disease. Int. J. Biol. Sci. 2018, 14, 633–643.
- Iyengar, A.; Maron, J.L. Detecting Infection in Neonates: Promises and Challenges of a Salivary Approach. Clin. Ther. 2015, 37, 523–528.
- Khanna, P.; Johnson, K.; Maron, J. Optimal Reference Genes for RT-QPCR Normalization in the Newborn. Biotech. Histochem. 2017, 92, 459–466.
- Carvalho, C.G.; Procianoy, R.S.; Neto, E.C.; Silveira, R.C. Preterm Neonates with Respiratory Distress Syndrome: Ventilator-Induced Lung Injury and Oxidative Stress. J. Immunol. Res. 2018, 2018, 6963754.
- Cadamuro, J.; Ibarz, M.; Cornes, M.; Nybo, M.; Haschke-Becher, E.; von Meyer, A.; Lippi, G.; Simundic, A.-M. Managing Inappropriate Utilization of Laboratory Resources. Diagnosis 2019, 6, 5–13.
- Mrazek, C.; Simundic, A.-M.; Salinas, M.; von Meyer, A.; Cornes, M.; Bauçà, J.M.; Nybo, M.; Lippi, G.; Haschke-Becher, E.; Keppel, M.H.; et al. Inappropriate Use of Laboratory Tests: How Availability Triggers Demand—Examples across Europe. Clin. Chim. Acta 2020, 505, 100–107.
- Thébaud, B.; Goss, K.N.; Laughon, M.; Whitsett, J.A.; Abman, S.H.; Steinhorn, R.H.; Aschner, J.L.; Davis, P.G.; McGrath-Morrow, S.A.; Soll, R.F.; et al. Bronchopulmonary Dysplasia. Nat. Rev. Dis. Prim. 2019, 5, 78.
- Hanna, N.; Kiefer, D. A Translational View of Biomarkers in Preterm Labor. Am J Reprod Immunol 2012, 67, 268–272.
- Singh, H.; Cho, S.J.; Gupta, S.; Kaur, R.; Sunidhi, S.; Saluja, S.; Pandey, A.K.; Bennett, M.V.; Lee, H.C.; Das, R.; et al. Designing a Bed-Side System for Predicting Length of Stay in a Neonatal Intensive Care Unit. Sci. Rep. 2021, 11, 3342.
- Ding, L.; Wang, H.; Geng, H.; Cui, N.; Huang, F.; Zhu, X.; Zhu, X. Prediction of Bronchopulmonary Dysplasia in Preterm Infants Using Postnatal Risk Factors. Front. Pediatr. 2020, 8, 349.
- Lure, A.C.; Du, X.; Black, E.W.; Irons, R.; Lemas, D.J.; Taylor, J.A.; Lavilla, O.; de la Cruz, D.; Neu, J. Using Machine Learning Analysis to Assist in Differentiating between Necrotizing Enterocolitis and Spontaneous Intestinal Perforation: A Novel Predictive Analytic Tool. J. Pediatr. Surg. 2021, 56, 1703–1710.
- López-Martínez, F.; Núñez-Valdez, E.R.; Gomez, J.L.; García-Díaz, V. A Neural Network Approach to Predict Early Neonatal Sepsis. Comput. Electr. Eng. 2019, 76, 379–388.
- Gehle, D.B.; Chapman, A.; Gregoski, M.; Brunswick, M.; Anderson, E.; Ramakrishnan, V.; Muhammad, L.N.; Head, W.; Lesher, A.P.; Ryan, R.M. A Predictive Model for Preterm Babies Born < 30 Weeks Gestational Age Who Will Not Attain Full Oral Feedings. J. Perinatol. 2022, 42, 126–131.
More
Information
Subjects:
Pediatrics
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Entry Collection:
Extraction Techniques in Sample Preparation
Revisions:
2 times
(View History)
Update Date:
15 Feb 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




