
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sara Domingues | -- | 2233 | 2023-02-06 11:28:30 | | | |
| 2 | Lindsay Dong | -5 word(s) | 2228 | 2023-02-07 03:43:28 | | |
Video Upload Options
Gram-negative bacteria are resistant to many commercialized antibiotics. The outer membrane (OM) of Gram-negative bacteria prevents the entry of such antibiotics. Outer membrane vesicles (OMV) are naturally released from the OM of Gram-negative bacteria for a range of purposes, including competition with other bacteria. OMV may carry, as part of the membrane or lumen, molecules with antibacterial activity. Such OMV can be exposed to and can fuse with the cell surface of different bacterial species.
1. Introduction
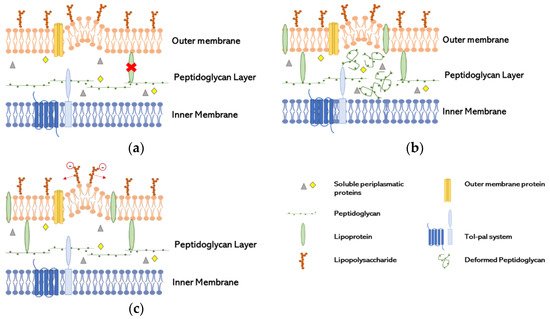
2. OMV Biogenesis in Gram-Negative Bacteria
-
LPP links deficiency (Figure 1a): the presence of LPP in the unbound form has been found in OMV, indicating that the covalent links were broken, or their distribution was not homogenous, since the conversion of free-form LPP into bound form is reversible [21]. These characteristics seems to be induced by the non-proportional growth of the OM compared with the PG layer [22]. The relation of the lack of link between OmpA and PG has been proven to be essential to the production of OMV in Salmonella spp. [23].
-
Increase of misfolded PG (Figure 1b): Autolysins have a role in cleaving the covalent links of PG, resulting in cell wall remodeling. The lack of these enzymes increases the amount of peptides in periplasmatic space and other components leading to turgor pressure and therefore to OMV formation. Several studies explore the lack of autolysins to increase the concentration of proteins in the periplasmatic space and therefore converge to this model [24][25].
-
Repulsion of negatively charged LPS (Figure 1c): A study suggested that the repulsion of negatively charge B-band LPS in cells exposed to gentamicin, with great affinity to LPS, induce the release of OMV as a way of antibiotic resistance in which gentamicin was incorporated into OMV. That repulsion increased the production of vesicles in P. aeruginosa [6].
3. Antimicrobial Activity of OMV
The OMV antimicrobial activity is appealing for treatment purposes, especially against Gram-negative bacteria since it allows bridging with its cell wall. During OMV biogenesis, some molecules with antibacterial activity will be naturally included in the vesicle lumen. At the same time, several environmental inductors can enhance or contribute to the inclusion of those molecules into the OMV. In both cases, OMV can act as an antimicrobial agent.
3.1. OMV with Natural Antimicrobial Activity Cargo
3.2. OMV with Loaded Antimicrobial Cargo
There are two main ways to incorporate antibiotics into OMV: the passive loading, where the addition of the antibiotics during bacterial growth is enough to produce antibiotic-carrying OMV (aOMV), and the active loading approach, where the antibiotics are forced to enter or coat the OMV or OM of bacteria, so it can be part of the produced OMV.
3.2.1. Passive Loading
3.2.2. Active Loading
4. Conclusions
References
- Garaeva, L.; Kamyshinsky, R.; Kil, Y.; Varfolomeeva, E.; Verlov, N.; Komarova, E.; Garmay, Y.; Landa, S.; Burdakov, V.; Myasnikov, A.; et al. Delivery of Functional Exogenous Proteins by Plant-Derived Vesicles to Human Cells in Vitro. Sci. Rep. 2021, 11, 6489.
- Fiocca, R.; Necchi, V.; Sommi, P.; Ricci, V.; Telford, J.; Cover, T.L.; Solcia, E. Release of Helicobacter Pylori Vacuolating Cytotoxin by Both a Specific Secretion Pathway and Budding of Outer Membrane Vesicles. Uptake of Released Toxin and Vesicles by Gastric Epithelium. J. Pathol. 1999, 188, 220–226.
- Marchant, P.; Carreño, A.; Vivanco, E.; Silva, A.; Nevermann, J.; Otero, C.; Araya, E.; Gil, F.; Calderón, I.L.; Fuentes, J.A. “One for All”: Functional Transfer of OMV-Mediated Polymyxin B Resistance From Salmonella Enterica Sv. Typhi ΔtolR and ΔdegS to Susceptible Bacteria. Front. Microbiol. 2021, 12, 1–15.
- Shima, T.; Muraoka, T.; Hamada, T.; Morita, M.; Takagi, M.; Fukuoka, H.; Inoue, Y.; Sagawa, T.; Ishijima, A.; Omata, Y.; et al. Micrometer-Size Vesicle Formation Triggered by UV Light. Langmuir 2014, 30, 7289–7295.
- Redeker, C.; Briscoe, W.H. Interactions between Mutant Bacterial Lipopolysaccharide (LPS-Ra) Surface Layers: Surface Vesicles, Membrane Fusion, and Effect of Ca2+ and Temperature. Langmuir 2019, 35, 15739–15750.
- Kadurugamuwa, J.L.; Beveridge, T.J. Virulence Factors Are Released from Pseudomonas Aeruginosa in Association with Membrane Vesicles during Normal Growth and Exposure to Gentamicin: A Novel Mechanism of Enzyme Secretion. J. Bacteriol. 1995, 177, 3998–4008.
- Chatterjee, S.; Mondal, A.; Mitra, S.; Basu, S. Acinetobacter Baumannii Transfers the BlaNDM-1 Gene via Outer Membrane Vesicles. J. Antimicrob. Chemother. 2017, 72, 2201–2207.
- Lin, J.; Zhang, W.; Cheng, J.; Yang, X.; Zhu, K.; Wang, Y.; Wei, G.; Qian, P.-Y.; Luo, Z.-Q.; Shen, X. A Pseudomonas T6SS Effector Recruits PQS-Containing Outer Membrane Vesicles for Iron Acquisition. Nat. Commun. 2017, 8, 14888.
- Fuhrmann, G.; Serio, A.; Mazo, M.; Nair, R.; Stevens, M.M. Active Loading into Extracellular Vesicles Significantly Improves the Cellular Uptake and Photodynamic Effect of Porphyrins. J. Control. Release 2015, 205, 35–44.
- Michel, L.V.; Gallardo, L.; Konovalova, A.; Bauer, M.; Jackson, N.; Zavorin, M.; McNamara, C.; Pierce, J.; Cheng, S.; Snyder, E.; et al. Ampicillin Triggers the Release of Pal in Toxic Vesicles from Escherichia Coli. Int. J. Antimicrob. Agents 2020, 56, 106163.
- Marion, C.R.; Lee, J.; Sharma, L.; Park, K.; Lee, C.; Liu, W.; Liu, P.; Feng, J.; Gho, Y.S.; Dela Cruz, C.S. Toll-Like Receptors 2 and 4 Modulate Pulmonary Inflammation and Host Factors Mediated by Outer Membrane Vesicles Derived from Acinetobacter Baumannii. Infect. Immun. 2019, 87, e00243-19.
- Kadurugamuwa, J.L.; Beveridge, T.J. Bacteriolytic Effect of Membrane Vesicles from Pseudomonas Aeruginosa on Other Bacteria Including Pathogens: Conceptually New Antibiotics. J. Bacteriol. 1996, 178, 2767–2774.
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and Origins of Bacterial Membrane Vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24.
- Domingues, S.; Nielsen, K.M. Membrane Vesicles and Horizontal Gene Transfer in Prokaryotes. Curr. Opin. Microbiol. 2017, 38, 16–21.
- Domingues, S.; Rosário, N.; Ben Cheikh, H.; Da Silva, G.J. ISAba1 and Tn6168 Acquisition by Natural Transformation Leads to Third-Generation Cephalosporins Resistance in Acinetobacter Baumannii. Infect. Genet. Evol. 2018, 63, 13–16.
- Park, J.; Kim, M.; Shin, B.; Kang, M.; Yang, J.; Lee, T.K.; Park, W. A Novel Decoy Strategy for Polymyxin Resistance in Acinetobacter Baumannii. Elife 2021, 10, 1–29.
- Collins, S.M.; Brown, A.C. Bacterial Outer Membrane Vesicles as Antibiotic Delivery Vehicles. Front. Immunol. 2021, 12, 733064.
- Schwechheimer, C.; Sullivan, C.J.; Kuehn, M.J. Envelope Control of Outer Membrane Vesicle Production in Gram-Negative Bacteria. Biochemistry 2013, 52, 3031–3040.
- Avila-Calderón, E.D.; Ruiz-Palma, M.D.S.; Aguilera-Arreola, M.G.; Velázquez-Guadarrama, N.; Ruiz, E.A.; Gomez-Lunar, Z.; Witonsky, S.; Contreras-Rodríguez, A. Outer Membrane Vesicles of Gram-Negative Bacteria: An Outlook on Biogenesis. Front. Microbiol. 2021, 12, 557902.
- Lee, E.-Y.; Bang, J.Y.; Park, G.W.; Choi, D.-S.; Kang, J.S.; Kim, H.-J.; Park, K.-S.; Lee, J.-O.; Kim, Y.-K.; Kwon, K.-H.; et al. Global Proteomic Profiling of Native Outer Membrane Vesicles Derived from Escherichia Coli. Proteomics 2007, 7, 3143–3153.
- Hoekstra, D.; van der Laan, J.W.; de Leij, L.; Witholt, B. Release of Outer Membrane Fragments from Normally Growing Escherichia Coli. Biochim. Biophys. Acta 1976, 455, 889–899.
- Wensink, J.; Witholt, B. Outer-Membrane Vesicles Released by Normally Growing Escherichia Coli Contain Very Little Lipoprotein. Eur. J. Biochem. 1981, 116, 331–335.
- Deatherage, B.L.; Lara, J.C.; Bergsbaken, T.; Rassoulian Barrett, S.L.; Lara, S.; Cookson, B.T. Biogenesis of Bacterial Membrane Vesicles. Mol. Microbiol. 2009, 72, 1395–1407.
- Hayashi, J.; Hamada, N.; Kuramitsu, H.K. The Autolysin of Porphyromonas Gingivalis Is Involved in Outer Membrane Vesicle Release. FEMS Microbiol. Lett. 2002, 216, 217–222.
- Zhou, L.; Srisatjaluk, R.; Justus, D.E.; Doyle, R.J. On the Origin of Membrane Vesicles in Gram-Negative Bacteria. FEMS Microbiol. Lett. 1998, 163, 223–228.
- Li, Z.; Clarke, A.J.; Beveridge, T.J. A Major Autolysin of Pseudomonas Aeruginosa: Subcellular Distribution, Potential Role in Cell Growth and Division and Secretion in Surface Membrane Vesicles. J. Bacteriol. 1996, 178, 2479–2488.
- Evans, A.G.L.; Davey, H.M.; Cookson, A.; Currinn, H.; Cooke-Fox, G.; Stanczyk, P.J.; Whitworth, D.E. Predatory Activity of Myxococcus Xanthus Outer-Membrane Vesicles and Properties of Their Hydrolase Cargo. Microbiology 2012, 158, 2742–2752.
- HOSODA, J.; NOMURA, M. Nature of the Primary Action of the Autolysin of Bacillus Subtilis. J. Bacteriol. 1956, 72, 573–581.
- Ghuysen, J.M. Use of Bacteriolytic Enzymes in Determination of Wall Structure and Their Role in Cell Metabolism. Bacteriol. Rev. 1968, 32, 425–464.
- Oshida, T.; Sugai, M.; Komatsuzawa, H.; Hong, Y.M.; Suginaka, H.; Tomasz, A. A Staphylococcus Aureus Autolysin That Has an N-Acetylmuramoyl-L-Alanine Amidase Domain and an Endo-Beta-N-Acetylglucosaminidase Domain: Cloning, Sequence Analysis, and Characterization. Proc. Natl. Acad. Sci. USA 1995, 92, 285–289.
- Hodak, H.; Galán, J.E. A Salmonella Typhi Homologue of Bacteriophage Muramidases Controls Typhoid Toxin Secretion. EMBO Rep. 2013, 14, 95–102.
- Scheurwater, E.; Reid, C.W.; Clarke, A.J. Lytic Transglycosylases: Bacterial Space-Making Autolysins. Int. J. Biochem. Cell Biol. 2008, 40, 586–591.
- Ishida, S.T.; Ishida, T. Zinc-Induced Immune Anti-Bacterial Vaccine Activity in Bacterial Cell Walls against Gram-Positive and Gram-Negative Bacteria. Proc. ARC J. Immunol. Vaccines 2019, 4, 10–19.
- Yamada, S.; Sugai, M.; Komatsuzawa, H.; Nakashima, S.; Oshida, T.; Matsumoto, A.; Suginaka, H. An Autolysin Ring Associated with Cell Separation of Staphylococcus Aureus. J. Bacteriol. 1996, 178, 1565–1571.
- Li, Z.; Clarke, A.J.; Beveridge, T.J. Gram-Negative Bacteria Produce Membrane Vesicles Which Are Capable of Killing Other Bacteria. J. Bacteriol. 1998, 180, 5478–5483.
- Kudryakova, I.V.; Suzina, N.E.; Vasilyeva, N.V. Biogenesis of Lysobacter Sp. XL1 Vesicles. FEMS Microbiol. Lett. 2015, 362, fnv137.
- Chen, S.; Yang, D.; Wen, Y.; Jiang, Z.; Zhang, L.; Jiang, J.; Chen, Y.; Hu, T.; Wang, Q.; Zhang, Y.; et al. Dysregulated Hemolysin Liberates Bacterial Outer Membrane Vesicles for Cytosolic Lipopolysaccharide Sensing. PLoS Pathog. 2018, 14, e1007240.
- Hood, R.D.; Singh, P.; Hsu, F.; Güvener, T.; Carl, M.A.; Trinidad, R.R.S.; Silverman, J.M.; Ohlson, B.B.; Hicks, K.G.; Plemel, R.L.; et al. A Type VI Secretion System of Pseudomonas Aeruginosa Targets a Toxin to Bacteria. Cell Host Microbe 2010, 7, 25–37.
- Russell, A.B.; Hood, R.D.; Bui, N.K.; LeRoux, M.; Vollmer, W.; Mougous, J.D. Type VI Secretion Delivers Bacteriolytic Effectors to Target Cells. Nature 2011, 475, 343–347.
- Nikaido, H.; Vaara, M. Molecular Basis of Bacterial Outer Membrane Permeability. Microbiol. Rev. 1985, 49, 1–32.
- Blokhina, S.V.; Sharapova, A.V.; Ol’khovich, M.V.; Volkova, T.V.; Perlovich, G.L. Solubility, Lipophilicity and Membrane Permeability of Some Fluoroquinolone Antimicrobials. Eur. J. Pharm. Sci. 2016, 93, 29–37.
- Huang, W.; Zhang, Q.; Li, W.; Yuan, M.; Zhou, J.; Hua, L.; Chen, Y.; Ye, C.; Ma, Y. Development of Novel Nanoantibiotics Using an Outer Membrane Vesicle-Based Drug Efflux Mechanism. J. Control. Release 2020, 317, 1–22.
- Weaver, J.C. Electroporation Theory. Concepts and Mechanisms. Methods Mol. Biol. 1995, 47, 1–26.
- Stankevic, V.; Simonis, P.; Zurauskiene, N.; Stirke, A.; Dervinis, A.; Bleizgys, V.; Kersulis, S.; Balevicius, S. Compact Square-Wave Pulse Electroporator with Controlled Electroporation Efficiency and Cell Viability. Symmetry 2020, 12, 412.
- Gujrati, V.; Kim, S.; Kim, S.-H.; Min, J.J.; Choy, H.E.; Kim, S.C.; Jon, S. Bioengineered Bacterial Outer Membrane Vesicles as Cell-Specific Drug-Delivery Vehicles for Cancer Therapy. ACS Nano 2014, 8, 1525–1537.
- Pick, U. Liposomes with a Large Trapping Capacity Prepared by Freezing and Thawing of Sonicated Phospholipid Mixtures. Arch. Biochem. Biophys. 1981, 212, 186–194.
- Lamichhane, T.N.; Jeyaram, A.; Patel, D.B.; Parajuli, B.; Livingston, N.K.; Arumugasaamy, N.; Schardt, J.S.; Jay, S.M. Oncogene Knockdown via Active Loading of Small RNAs into Extracellular Vesicles by Sonication. Cell. Mol. Bioeng. 2016, 9, 315–324.
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of Exosome-Encapsulated Paclitaxel to Overcome MDR in Cancer Cells. Nanomedicine 2016, 12, 655–664.
- McCaig, W.D.; Loving, C.L.; Hughes, H.R.; Brockmeier, S.L. Characterization and Vaccine Potential of Outer Membrane Vesicles Produced by Haemophilus Parasuis. PLoS ONE 2016, 11, e0149132.
- Li, K.; Yuan, X.-X.; Sun, H.-M.; Zhao, L.-S.; Tang, R.; Chen, Z.-H.; Qin, Q.-L.; Chen, X.-L.; Zhang, Y.-Z.; Su, H.-N. Atomic Force Microscopy of Side Wall and Septa Peptidoglycan From Bacillus Subtilis Reveals an Architectural Remodeling During Growth. Front. Microbiol. 2018, 9, 620.
- Foladori, P.; Laura, B.; Gianni, A.; Giuliano, Z. Effects of Sonication on Bacteria Viability in Wastewater Treatment Plants Evaluated by Flow Cytometry--Fecal Indicators, Wastewater and Activated Sludge. Water Res. 2007, 41, 235–243.
- Gomes, A.; van Oosten, M.; Bijker, K.L.B.; Boiten, K.E.; Salomon, E.N.; Rosema, S.; Rossen, J.W.A.; Natour, E.; Douglas, Y.L.; Kampinga, G.A.; et al. Sonication of Heart Valves Detects More Bacteria in Infective Endocarditis. Sci. Rep. 2018, 8, 12967.
- Zhang, Y.-F.; Shi, J.-B.; Li, C. Small Extracellular Vesicle Loading Systems in Cancer Therapy: Current Status and the Way Forward. Cytotherapy 2019, 21, 1122–1136.
- Gayán, E.; Govers, S.K.; Aertsen, A. Impact of High Hydrostatic Pressure on Bacterial Proteostasis. Biophys. Chem. 2017, 231, 3–9.
- Sharif, E.; Eftekhari, Z.; Mohit, E. The Effect of Growth Stage and Isolation Method on Properties of ClearColiTM Outer Membrane Vesicles (OMVs). Curr. Microbiol. 2021, 78, 1602–1614.




