Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Carmen Rodríguez García | -- | 2105 | 2023-02-03 13:29:11 | | | |
| 2 | Lindsay Dong | Meta information modification | 2105 | 2023-02-06 03:41:56 | | | | |
| 3 | Lindsay Dong | -1 word(s) | 2104 | 2023-02-06 03:43:14 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Rodríguez-García, C.; Gutiérrez-Santiago, F. Post-Translational Modifications in Colorectal Cancer. Encyclopedia. Available online: https://encyclopedia.pub/entry/40813 (accessed on 07 February 2026).
Rodríguez-García C, Gutiérrez-Santiago F. Post-Translational Modifications in Colorectal Cancer. Encyclopedia. Available at: https://encyclopedia.pub/entry/40813. Accessed February 07, 2026.
Rodríguez-García, Carmen, Francisco Gutiérrez-Santiago. "Post-Translational Modifications in Colorectal Cancer" Encyclopedia, https://encyclopedia.pub/entry/40813 (accessed February 07, 2026).
Rodríguez-García, C., & Gutiérrez-Santiago, F. (2023, February 03). Post-Translational Modifications in Colorectal Cancer. In Encyclopedia. https://encyclopedia.pub/entry/40813
Rodríguez-García, Carmen and Francisco Gutiérrez-Santiago. "Post-Translational Modifications in Colorectal Cancer." Encyclopedia. Web. 03 February, 2023.
Copy Citation
Colorectal cancer (CRC) is one of the most common cancers worldwide. Its main modifiable risk factors are diet, alcohol consumption, and smoking. Thus, the right approach through lifestyle changes may lead to its prevention. Although cancer is a multi-factorial process, the study of post-translational modifications (PTMs) of proteins associated with CRC has gained interest, as inappropriate modification is closely related to the activation of cell signalling pathways involved in carcinogenesis.
colorectal cancer
(CRC)
post-translational modifications
(PTMs)
1. Introduction
Colorectal cancer (CRC) is currently the second type of cancer with the highest mortality rate in the population according to Global Cancer Statistics 2020 [1]. Metastatic CRC has a poor prognosis, with less than a 15% of five-year survival rate [2]. Its carcinogenesis is a process of many years of development and some early life risk factors are important contributors [3]. Among them, cigarette smoking, obesity, and a sedentary lifestyle are closely related to CRC incidence [4][5]. However, its quickly increasing incidence is mainly due to lifestyle westernization associated with changes in dietary behaviour such as heavy alcohol consumption and diets rich in sugars, saturated fats, and red and processed meat [6]. Thus, some protective lifestyle factors against CRC include a diet rich in minerals and vitamins, dairy, dietary fibre, fish, vegetables, and fruits. An alternative strategy for CRC prevention is the use of a chemopreventive supplement providing greater individual exposure to some nutrients than can be obtained from the diet (such as phytochemicals) [7].
The pathogenesis of CRC is a complex multi-stage process which includes gut microbiota imbalances, cell DNA disruption, and carcinogenic signalling pathways activation [8]. The aetiology underlying the mechanism of action of specific nutrients in CRC has been mainly attributed to their anti-inflammatory and antioxidant properties, and their modulation of gut microbiota populations, maintaining gut homeostasis and regulating the host immune response [9][10]. However, their effects on epigenetic modulation associated with CRC pathogenesis remains unknown. There is increasing evidence that the disruption of epigenetic control over gene expression has an important role in carcinogenesis [11][12][13][14]. Together with non-coding RNAs and DNA methylation, histone and protein post-translational modifications (PTMs) have an important role in carcinogenesis and gene regulation [15][16]. PTMs occur once the mRNA has been translated into the protein sequence in the ribosomes and produce marginal chemical modifications to lipoproteins and native proteins. Among these modifications, PTMs may mark proteins for degradation, inhibit or promote interactions with other proteins, redirect cellular protein localization, and modify enzyme activity [17][18]. Most PTMs are reversible, so normal cells use them as a switch to control proliferating or quiescent cells [19]. The role of PTMs in the onset and progression of diseases such as cancer has been investigated. Their involvement in the process of carcinogenesis could be due to their function in processes such as the cell cycle, cell survival, and cell proliferation [20]. Therefore, PTM-focused analysis of enzyme phosphorylation and the involvement of protein kinases in cancer formation and progression have led to the use of PTM-based therapeutic approaches (i.e., tyrosine kinase inhibitors) [21][22]. Furthermore, in the case of CRC, PTMs develop key role-playing as a tight junction protein and regulate the epithelial barrier function [23][24]. Thus, PTMs may be essential to work with the external impact and could provide an excellent opportunity for intervention through feeding and promoting clinical strategies for CRC patients regarding predictive, preventive, and personalized medicine.
2. Post-Translational Modifications in Colorectal Cancer
PTMs are protein-specific modifications that control many physiological processes to ensure the dynamic and quick response of cells to intracellular and extracellular stimuli [25]. Any proteome protein may be modified post-translationally or during translation. These reversible modifications may alter not only the protein’s stability, conformation, and charge state, but also its function modulating its intracellular conformation, its interactions, and the life span of the target protein [26]. In some cases, PTMs are inadequate and modulate positively some signal transduction pathways that are involved in tumourigenesis regulation and cancer development [27]. To date, more than 450 unique protein modifications have been described, including ubiquitination, acylation, SUMOylation, methylation, and phosphorylation [28]. In the case of CRC, the most important modifications involved have been summarized below (Figure 1).
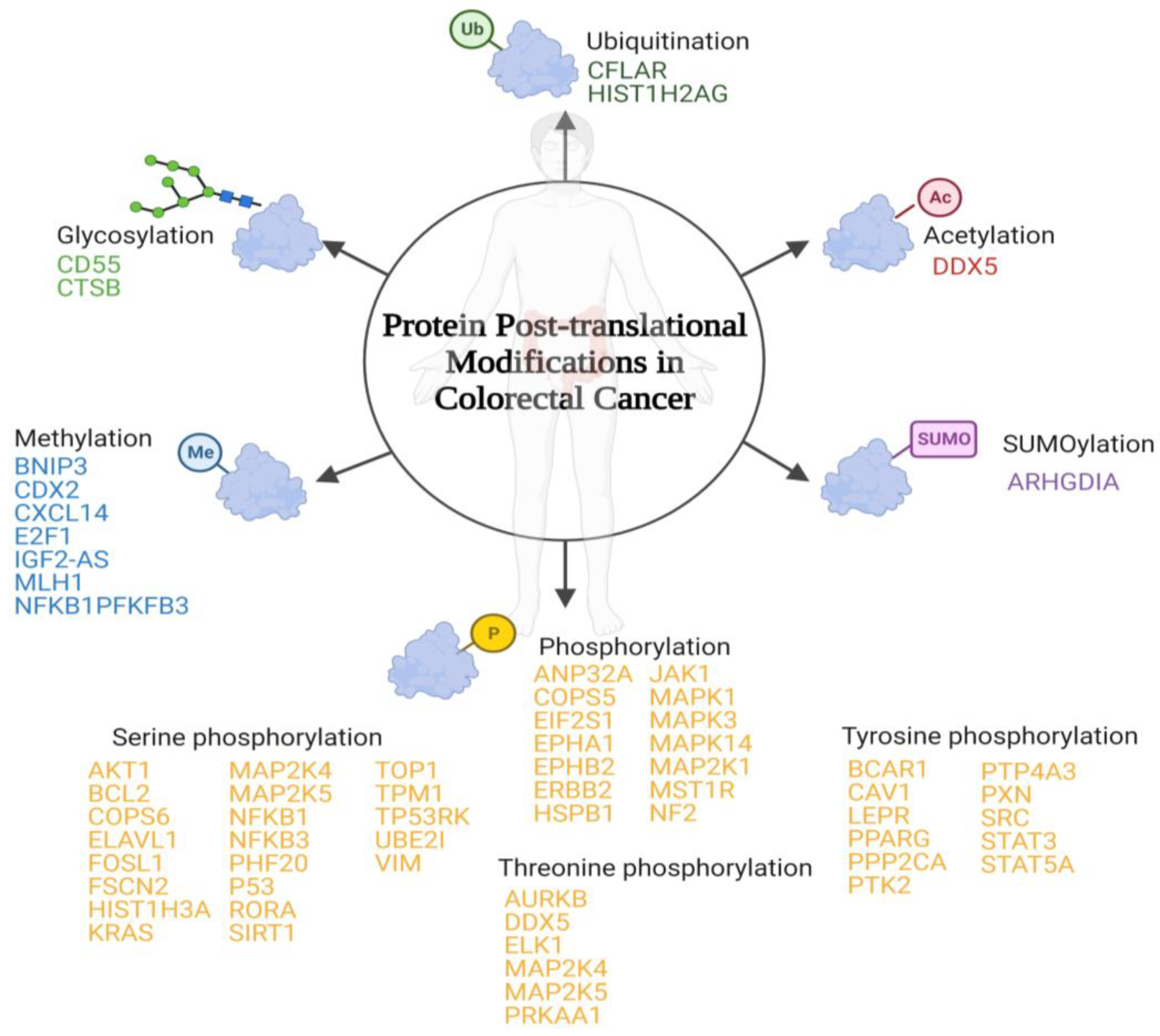
Figure 1. Schematic representation of the main post-translational modifications in colorectal cancer. Below each post-translational modification is a list of the identified proteins that suffer inappropriate post-translational modifications associated with colorectal cancer.
2.1. SUMOylation
Small ubiquitin-like modifiers (SUMO) are covalently attached to lysine residues [29]. The downregulated SUMOylation in lysine 138 of Rho GDP-dissociation inhibitor 1 has been observed in CRC cell lines. This protein is involved in Rho GTPases signalling regulation [30].
2.2. Glycosylation
A carbohydrate is attached to specific proteins. In mammals, there are two types: (1) O-glycosylation, where glycosyl groups are connected to tyrosine, hydroxylysine, serine, or threonine side chains with glycosidic linkages by glycosyltransferases, and (2) N-glycosylation, where glycosyl groups are connected to Asn side chains with amide linkages by oligosaccharyltransferase [31][32]. The upregulation of this PTM in complement decay-accelerating factor and cathepsin B has been identified in tumour tissue samples of CRC patients [33][34].
2.3. O-GlcNAcylation
There is a covalent attachment of N-acetylglucosamine residue O-linked to the hydroxyl group of threonine and serine residues of multiple cytosolic and nuclear proteins [35][36]. The upregulation of O-GlcNAcylation in ATP-dependent RNA helicase DDX5 has been associated with CRC in cell lines and murine models [37].
2.4. Ubiquitination
There is an attachment of ubiquitin molecules to the lysine residue of the substrate proteins. This process is based on an enzymatic cascade of ubiquitin-activating, ubiquitin-conjugating, and ubiquitin-ligase enzymes [38][39]. There have been two ubiquitination-susceptible proteins identified that are related to CRC: (1) caspase homolog that is an apoptosis regulator [40] and (2) histone H2A type 1 that is involved in chromosomal stability, DNA replication, and DNA repair [41].
2.5. Methylation
Methylation occurs mainly in arginine or lysine residues. One of the most biologically important roles of methylation is in histone modification [42]. Among the different proteins that suffer dysregulated post-translational methylation associated with CRC, the one that is involved in cell growth suppression has downregulated methylation (putative insulin-like growth factor 2 antisense gene protein) [43][44]. The other proteins identified have an upregulated methylation, among them are (1) BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 that is involved in apoptosis [45]; (2) homeobox protein CDX-2 that is involved in the transcriptional regulation of different genes expressed in the intestine [46]; (3) C-X-C motif chemokine 14 that is involved in immunoregulatory and inflammatory processes [47]; (4) transcription factor E2F1 that participates in the cell cycle [48]; (5) DNA mismatch repair protein Mlh1 that participates in DNA repair [49]; (6) nuclear factor NF-kappa-B p105 subunit that is a pleiotropic transcription factor involved in several signal transduction events which are initiated by stimuli such as oxidative stress or inflammation [50]; and (7) 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 that is an essential protein for cell cycle progression and apoptosis prevention [51].
2.6. Phosphorylation
Phosphorylation is the most prevalent and widely studied type of PTM. It is inversely regulated by phosphatases and protein kinases in the amino acids’ hydroxyl tyrosine, threonine, or serine [31][52]. In the case of CRC, inadequate PTMs have been identified in the following proteins: (1) acidic leucine-rich nuclear phosphoprotein 32 family member A that is involved in cell growth [53]; (2) COP9 signalosome complex subunit 5 that develops an important role in the degradation of cyclin-dependent kinase inhibitor [54]; (3) eukaryotic translation initiation factor 2 subunit 1 that is a translation initiation factor [55]; (4) ephrin type-A receptor 1 and ephrin type-B receptor 2 that are members of the ephrin receptor subfamily of the protein tyrosine kinase family [56]; (5) receptor tyrosine-protein kinase erbB-2 that is a member of the epidermal growth factor receptor family [57]; (6) heat shock protein beta-1 which plays an important role in cancer cells proliferation [58]; (7) tyrosine-protein kinase JAK1 that is a tyrosine kinase of the non-receptor type [59]; (8) mitogen-activated protein kinase 1, 3, and 14 that are serine/threonine kinases that are essential components of the MAP kinase signal transduction pathway [60][61][62][63]; (9) dual specificity mitogen-activated protein kinase kinase 1 which acts as an essential component of the MAP kinase signal transduction pathway [64]; (10) macrophage-stimulating protein receptor that is a tyrosine kinase receptor [65]; and (11) merlin that plays a pivotal role in tumour suppression through apoptosis promotion [66].
2.7. Serine Phosphorylation
Serine phosphorylation includes proto-oncogene c-Ak and Fos-related antigen 1 that regulates many processes including proliferation cell survival, growth, and angiogenesis [67][68]; apoptosis regulator Bcl-2 that is a regulator of apoptosis [69]; COP9 signalosome complex subunit 6 which is a component of the COP9 signalosome complex [70]; ELAV-like protein 1 that stabilizes mRNAs and regulates gene expression [71]; fascin-2 that acts as an actin bundling protein [72]; histone H3.1 which plays a central role in transcription regulation and DNA repair [73]; Kirsten rat sarcoma virus which is involved in the propagation of growth factors [74]; MAP kinase kinase 4 and 5 that are dual specificity protein kinase which act as an essential component of the MAP kinase signal transduction pathway [75][76]; NFKB1 and NFKB3 which are pleiotropic transcription factors involved in several signal transduction [77][78]; PHD finger protein 20 that contributes to p53 stabilization after DNA damage [79]; cellular tumour antigen p53 that acts as a tumour suppressor [80]; nuclear receptor ROR-alpha which is a key regulator of glucose metabolism [81]; sirtuin 1 that is an intracellular regulatory protein [82]; DNA topoisomerase 1 that releases the supercoiling tension of DNA introduced during the DNA replication [83]; tropomyosin-1 which is a member of the tropomyosin family of highly conserved proteins [84]; TP53-regulating kinase which is a protein kinase that phosphorylates ‘Ser-15’ of p53/TP53 protein [85]; SUMO-protein ligase that is essential for nuclear architecture and chromosome segregation [86]; and vimentin which is responsible for maintaining cell shape and stabilizing cytoskeletal interactions [87].
2.8. Threonine Phosphorylation
Threonine phosphorylation includes Aurora kinase B which is a serine/threonine-protein kinase component of the chromosomal passenger complex [88]; probable ATP-dependent RNA helicase DDX5 which is involved in the alternative regulation of pre-mRNA splicing [89]; ETS domain-containing protein Elk-1 which is a transcription factor that binds to purine-rich DNA sequences [90]; dual specificity mitogen-activated protein kinase kinase 4 which is an essential component of the MAP kinase signal transduction pathway [75]; MAP kinase kinase 5 that acts as a scaffold for the formation of a ternary MAP3K2/MAP3K3-MAP3K5-MAPK7 signalling complex [76]; and 5′-AMP-activated protein kinase catalytic subunit alpha-1 which is the catalytic subunit of AMP-activated protein kinase that plays a key role in regulating cellular energy metabolism [91].
2.9. Tyrosine Phosphorylation
Tyrosine phosphorylation includes breast cancer anti-estrogen resistance protein 1 which plays a central role in cell adhesion [92][93]; caveolin-1 that act as a scaffolding protein within caveolar membranes [94]; leptin receptor that mediates leptin central and peripheral effects [95]; peroxisome proliferator-activated receptor gamma that is a nuclear receptor [96]; serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform which is the major phosphatase for microtubule-associated proteins [97]; focal adhesion kinase 1 which is a non-receptor protein-tyrosine kinase that plays an essential role in regulating cell migration and apoptosis [98][99]; protein tyrosine phosphatase type IVA 3 that stimulates progression from G1 into S phase during mitosis [100]; paxillin which is a cytoskeletal protein involved in actin-membrane attachment at sites of cell adhesion to the extracellular matrix [101]; proto-oncogene tyrosine-protein kinase Src that is a non-receptor protein tyrosine kinase [102]; signal transducer and activator of transcription 3 which mediates cellular responses to interleukins and other growth factors [103][104]; and signal transducer and activator of transcription 5A that is involved in signal transduction and activation of transcription [105].
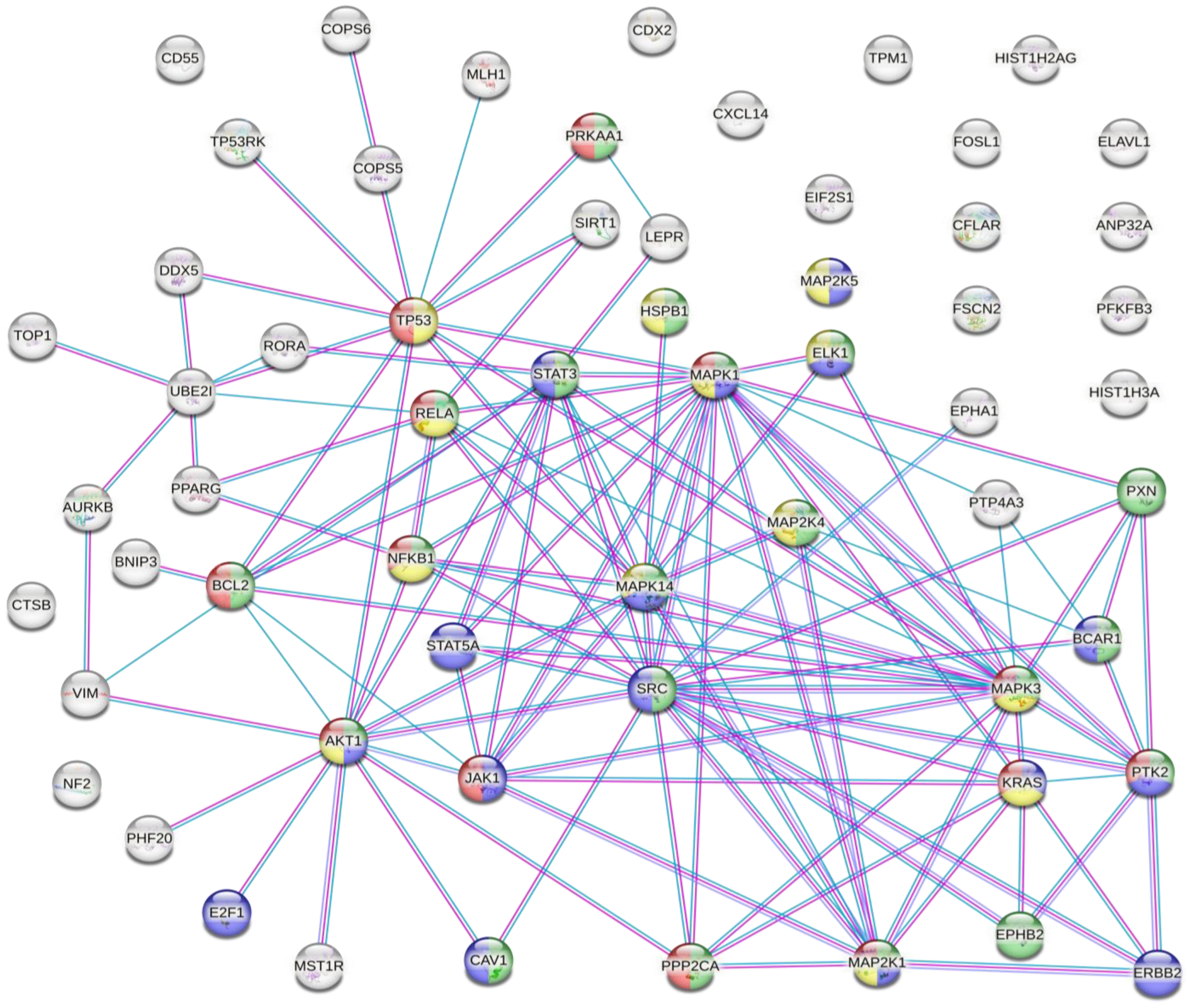
3. Relationship between Post-Translational Modifications Associated with Colorectal Cancer
The results of the analysis showed that there were several interactions between some of the proteins susceptible to inappropriate PTMs associated with CRC (Figure 2).

Figure 2. Protein–protein interaction network. Coloured nodes in green: proteins involved in the VEGFA-VEGFR2 signalling pathway. Coloured nodes in blue: proteins involved in the EGF-EFGR signalling pathway. Coloured nodes in red: proteins involved in the MAPK signalling pathway. Coloured nodes in yellow: proteins involved in the PI3K-Akt signalling pathway. Coloured nodes in grey: proteins that are not involved in any of the signalling pathways mentioned above. Edges represent protein–protein associations. Pink line: association experimentally determined. Blue line: association determined from curated databases. Purple line: protein homology.
This analysis showed that there were strong interactions between TP53, AKT1, STAT3, STAT5A, JAK1, MAPK1, MAPK14, MAP2K1, and SRC. In fact, this network had significantly more interactions than expected, which means that proteins have more interactions among themselves than what would be expected from a random set of proteins, demonstrating that the proteins may be partially biologically connected as a group. This group of proteins is mainly involved in the PI3K-Akt, EGF-EFGR, MAPK, and VEGFA-VEGFR2 signalling pathways. On the one hand, PI3K-Akt is the classical signalling pathway involved in glucose metabolism that promotes cancer metabolic reprogramming by elevation of aerobic glycolysis (known as the “Warburg effect”) [106][107]. Both EGF-EGFR and MAPK signalling pathways are involved in proliferation, differentiation, and apoptosis. Its regulation in cancer cells allows the maintenance of proliferative signalling, promoting cancer cell survival [52][108]. On the other hand, VEGF and its receptors (such as VEGFR2) develop an important role in tumour-associated angiogenesis. This process is essential for tumour progression because it favours oxygen and nutrient uptake by cancer cells [109][110]. Therefore, the main PTMs identified in CRC are involved in cancer progression and cancer cell survival.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164.
- Hughes, L.A.E.; van den Brandt, P.A.; Goldbohm, R.A.; de Goeij, A.F.P.M.; de Bruïne, A.P.; van Engeland, M.; Weijenberg, M.P. Childhood and Adolescent Energy Restriction and Subsequent Colorectal Cancer Risk: Results from the Netherlands Cohort Study. Int. J. Epidemiol. 2010, 39, 1333–1344.
- Dacrema, M.; Ali, A.; Ullah, H.; Khan, A.; di Minno, A.; Xiao, J.; Martins, A.M.C.; Daglia, M. Spice-Derived Bioactive Compounds Confer Colorectal Cancer Prevention via Modulation of Gut Microbiota. Cancers 2022, 14, 5682.
- Nimptsch, K.; Wu, K. Is Timing Important? The Role of Diet and Lifestyle during Early Life on Colorectal Neoplasia. Curr. Color. Cancer Rep. 2018, 14, 1–11.
- Masdor, N.A.; Mohammed Nawi, A.; Hod, R.; Wong, Z.; Makpol, S.; Chin, S.-F. The Link between Food Environment and Colorectal Cancer: A Systematic Review. Nutrients 2022, 14, 3954.
- Kim, S.H.; Moon, J.Y.; Lim, Y.J. Dietary Intervention for Preventing Colorectal Cancer: A Practical Guide for Physicians. J. Cancer Prev. 2022, 27, 139–146.
- Chen, M.; Lin, W.; Li, N.; Wang, Q.; Zhu, S.; Zeng, A.; Song, L. Therapeutic Approaches to Colorectal Cancer via Strategies Based on Modulation of Gut Microbiota. Front. Microbiol. 2022, 13, 945533.
- O’keefe, S.J.D. Diet, Microorganisms and Their Metabolites, and Colon Cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706.
- Reddy, B.S. Diet and Colon Cancer: Evidence from Human and Animal Model Studies. In Diet, Nutrition, and Cancer: A Critical Evaluation; CRC Press: Boca Raton, FL, USA, 2018; pp. 47–66. ISBN 1351071408.
- Ullah, A.; Ullah, N.; Nawaz, T.; Aziz, T. Molecular Mechanisms of Sanguinarine in Cancer Prevention and Treatment. Anticancer Agents Med. Chem. 2022, 22.
- Iqbal, H.; Menaa, F.; Khan, N.; Razzaq, A.; Khan, Z.; Ullah, K.; Kamal, R.; Sohail, M.; Thiripuranathar, G.; Uzair, B.; et al. Two Promising Anti-Cancer Compounds, 2-Hydroxycinnaldehyde and 2-Benzoyloxycinnamaldehyde: Where Do We Stand? Comb. Chem. High. Throughput Screen. 2022, 25, 808–818.
- Su, Q.; Fan, M.; Wang, J.; Ullah, A.; Ghauri, M.A.; Dai, B.; Zhan, Y.; Zhang, D.; Zhang, Y. Sanguinarine Inhibits Epithelial–Mesenchymal Transition via Targeting HIF-1α/TGF-β Feed-Forward Loop in Hepatocellular Carcinoma. Cell Death Dis. 2019, 10, 939.
- Ullah, A.; Leong, S.W.; Wang, J.; Wu, Q.; Ghauri, M.A.; Sarwar, A.; Su, Q.; Zhang, Y. Cephalomannine Inhibits Hypoxia-Induced Cellular Function via the Suppression of APEX1/HIF-1α Interaction in Lung Cancer. Cell Death Dis. 2021, 12, 490.
- Hong, X.; Huang, H.; Qiu, X.; Ding, Z.; Feng, X.; Zhu, Y.; Zhuo, H.; Hou, J.; Zhao, J.; Cai, W. Targeting Posttranslational Modifications of RIOK1 Inhibits the Progression of Colorectal and Gastric Cancers. eLife 2018, 7, e29511.
- Prieto, P.; Jaén, R.I.; Calle, D.; Gómez-Serrano, M.; Núñez, E.; Fernández-Velasco, M.; Martín-Sanz, P.; Alonso, S.; Vázquez, J.; Cerdán, S. Interplay between Post-Translational Cyclooxygenase-2 Modifications and the Metabolic and Proteomic Profile in a Colorectal Cancer Cohort. World J. Gastroenterol. 2019, 25, 433.
- Das, T.; Shin, S.C.; Song, E.J.; Kim, E.E. Regulation of Deubiquitinating Enzymes by Post-Translational Modifications. Int. J. Mol. Sci. 2020, 21, 4028.
- Kuwahara, H.; Nishizaki, M.; Kanazawa, H. Nuclear Localization Signal and Phosphorylation of Serine350 Specify Intracellular Localization of DRAK2. J. Biochem. 2008, 143, 349–358.
- Chen, L.; Liu, S.; Tao, Y. Regulating Tumor Suppressor Genes: Post-Translational Modifications. Signal Transduct. Target. Ther. 2020, 5, 90.
- Carter, A.M.; Tan, C.; Pozo, K.; Telange, R.; Molinaro, R.; Guo, A.; de Rosa, E.; Martinez, J.O.; Zhang, S.; Kumar, N. Phosphoprotein-Based Biomarkers as Predictors for Cancer Therapy. Proc. Natl. Acad. Sci. USA 2020, 117, 18401–18411.
- Kwon, Y.W.; Jo, H.-S.; Bae, S.; Seo, Y.; Song, P.; Song, M.; Yoon, J.H. Application of Proteomics in Cancer: Recent Trends and Approaches for Biomarkers Discovery. Front. Med. (Lausanne) 2021, 8, 747333.
- Hermann, J.; Schurgers, L.; Jankowski, V. Identification and Characterization of Post-Translational Modifications: Clinical Implications. Mol. Asp. Med. 2022, 86, 101066.
- Dai Vu, L.; Gevaert, K.; de Smet, I. Protein Language: Post-Translational Modifications Talking to Each Other. Trends Plant Sci. 2018, 23, 1068–1080.
- Reiche, J.; Huber, O. Post-Translational Modifications of Tight Junction Transmembrane Proteins and Their Direct Effect on Barrier Function. Biochim. Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183330.
- Li, W.; Li, F.; Zhang, X.; Lin, H.-K.; Xu, C. Insights into the Post-Translational Modification and Its Emerging Role in Shaping the Tumor Microenvironment. Signal Transduct. Target. 2021, 6, 422.
- Han, Z.-J.; Feng, Y.-H.; Gu, B.-H.; Li, Y.-M.; Chen, H. The Post-Translational Modification, SUMOylation, and Cancer. Int. J. Oncol. 2018, 52, 1081–1094.
- Jaén, R.I.; Prieto, P.; Casado, M.; Martín-Sanz, P.; Boscá, L. Post-Translational Modifications of Prostaglandin-Endoperoxide Synthase 2 in Colorectal Cancer: An Update. World J. Gastroenterol. 2018, 24, 5454–5461.
- Liu, N.; Ling, R.; Tang, X.; Yu, Y.; Zhou, Y.; Chen, D. Post-Translational Modifications of BRD4: Therapeutic Targets for Tumor. Front. Oncol. 2022, 12, 847701.
- Celen, A.B.; Sahin, U. Sumoylation on Its 25th Anniversary: Mechanisms, Pathology, and Emerging Concepts. FEBS J. 2020, 287, 3110–3140.
- Yu, J.; Zhang, D.; Liu, J.; Li, J.; Yu, Y.; Wu, X.-R.; Huang, C. RhoGDI SUMOylation at Lys-138 Increases Its Binding Activity to Rho GTPase and Its Inhibiting Cancer Cell Motility. J. Biol. Chem. 2012, 287, 13752–13760.
- Wang, H.; Yang, L.; Liu, M.; Luo, J. Protein Post-Translational Modifications in the Regulation of Cancer Hallmarks. Cancer Gene Ther. 2022.
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate Protein Glycosylation: Diversity, Synthesis and Function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462.
- Nakagawa, M.; Mizuno, M.; Kawada, M.; Uesu, T.; Nasu, J.; Takeuchi, K.; Okada, H.; Endo, Y.; Fujita, T.; Tsuji, T. Polymorphic Expression of Decay-Accelerating Factor in Human Colorectal Cancer. J. Gastroenterol. Hepatol. 2001, 16, 184–189.
- Iacobuzio-Donahue, C.A.; Shuja, S.; Cai, J.; Peng, P.; Murnane, M.J. Elevations in Cathepsin B Protein Content and Enzyme Activity Occur Independently of Glycosylation during Colorectal Tumor Progression. J. Biol. Chem. 1997, 272, 29190–29199.
- Slawson, C.; Hart, G.W. O-GlcNAc Signalling: Implications for Cancer Cell Biology. Nat. Rev. Cancer 2011, 11, 678–684.
- Hart, G.W.; Copeland, R.J. Glycomics Hits the Big Time. Cell 2010, 143, 672–676.
- Wu, N.; Jiang, M.; Han, Y.; Liu, H.; Chu, Y.; Liu, H.; Cao, J.; Hou, Q.; Zhao, Y.; Xu, B.; et al. O-GlcNAcylation Promotes Colorectal Cancer Progression by Regulating Protein Stability and Potential Catcinogenic Function of DDX5. J. Cell Mol. Med. 2019, 23, 1354–1362.
- Bedford, L.; Lowe, J.; Dick, L.R.; Mayer, R.J.; Brownell, J.E. Ubiquitin-like Protein Conjugation and the Ubiquitin–Proteasome System as Drug Targets. Nat. Rev. Drug Discov. 2011, 10, 29–46.
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The Role of Ubiquitination in Tumorigenesis and Targeted Drug Discovery. Signal Transduct. Target. Ther. 2020, 5, 1–28.
- Song, X.; Kim, S.-Y.; Zhou, Z.; Lagasse, E.; Kwon, Y.T.; Lee, Y.J. Hyperthermia Enhances Mapatumumab-Induced Apoptotic Death through Ubiquitin-Mediated Degradation of Cellular FLIP(Long) in Human Colon Cancer Cells. Cell Death Dis. 2013, 4, e577.
- Yu, T.; Chen, X.; Zhang, W.; Colon, D.; Shi, J.; Napier, D.; Rychahou, P.; Lu, W.; Lee, E.Y.; Weiss, H.L.; et al. Regulation of the Potential Marker for Intestinal Cells, Bmi1, by β-Catenin and the Zinc Finger Protein KLF4. J. Biol. Chem. 2012, 287, 3760–3768.
- Zhang, X.; Wen, H.; Shi, X. Lysine Methylation: Beyond Histones. Acta Biochim. Biophys. Sin. (Shanghai) 2012, 44, 14–27.
- Huang, Z.; Su, G.; Bi, X.; Zhang, L.; Xu, Z.; Wang, G. Over-Expression of Long Non-Coding RNA Insulin-like Growth Factor 2-Antisense Suppressed Hepatocellular Carcinoma Cell Proliferation and Metastasis by Regulating the MicroRNA-520h/Cyclin-Dependent Kinase Inhibitor 1A Signaling Pathway. Bioengineered 2021, 12, 6952–6966.
- Li, T.; Chen, H.; Li, W.; Cui, J.; Wang, G.; Hu, X.; Hoffman, A.R.; Hu, J. Promoter Histone H3K27 Methylation in the Control of IGF2 Imprinting in Human Tumor Cell Lines. Hum. Mol. Genet. 2014, 23, 117–128.
- Matsunaga, Y.; Tamura, Y.; Takahashi, K.; Kitaoka, Y.; Takahashi, Y.; Hoshino, D.; Kadoguchi, T.; Hatta, H. Branched-chain Amino Acid Supplementation Suppresses the Detraining-induced Reduction of Mitochondrial Content in Mouse Skeletal Muscle. FASEB J. 2022, 36, e22628.
- Wang, Y.; Kou, Y.; Zhu, R.; Han, B.; Li, C.; Wang, H.; Wu, H.; Xia, T.; Che, X. CDX2 as a Predictive Biomarker Involved in Immunotherapy Response Suppresses Metastasis through EMT in Colorectal Cancer. Dis. Markers 2022, 2022, 9025668.
- Yang, X.-Y.; Ozawa, S.; Kato, Y.; Maehata, Y.; Izukuri, K.; Ikoma, T.; Kanamori, K.; Akasaka, T.; Suzuki, K.; Iwabuchi, H.; et al. C-X-C Motif Chemokine Ligand 14 Is a Unique Multifunctional Regulator of Tumor Progression. Int. J. Mol. Sci. 2019, 20, 1872.
- Dubrez, L. Regulation of E2F1 Transcription Factor by Ubiquitin Conjugation. Int. J. Mol. Sci. 2017, 18, 2188.
- Torres, K.A.; Calil, F.A.; Zhou, A.L.; DuPrie, M.L.; Putnam, C.D.; Kolodner, R.D. The Unstructured Linker of Mlh1 Contains a Motif Required for Endonuclease Function Which Is Mutated in Cancers. Proc. Natl. Acad. Sci. USA 2022, 119, e2212870119.
- Dobre, M.; Trandafir, B.; Milanesi, E.; Salvi, A.; Bucuroiu, I.A.; Vasilescu, C.; Niculae, A.M.; Herlea, V.; Hinescu, M.E.; Constantinescu, G. Molecular Profile of the NF-ΚB Signalling Pathway in Human Colorectal Cancer. J. Cell Mol. Med. 2022, 26, 5966–5975.
- Zhou, Z.; Plug, L.G.; Patente, T.A.; de Jonge-Muller, E.S.M.; Elmagd, A.A.; van der Meulen-de Jong, A.E.; Everts, B.; Barnhoorn, M.C.; Hawinkels, L.J.A.C. Increased Stromal PFKFB3-Mediated Glycolysis in Inflammatory Bowel Disease Contributes to Intestinal Inflammation. Front. Immunol. 2022, 13, 966067.
- Singh, V.; Ram, M.; Kumar, R.; Prasad, R.; Roy, B.K.; Singh, K.K. Phosphorylation: Implications in Cancer. Protein J. 2017, 36, 1–6.
- Yu, L.-G.; Packman, L.C.; Weldon, M.; Hamlett, J.; Rhodes, J.M. Protein Phosphatase 2A, a Negative Regulator of the ERK Signaling Pathway, Is Activated by Tyrosine Phosphorylation of Putative HLA Class II-Associated Protein I (PHAPI)/Pp32 in Response to the Antiproliferative Lectin, Jacalin. J. Biol. Chem. 2004, 279, 41377–41383.
- Nishimoto, A.; Kugimiya, N.; Hosoyama, T.; Enoki, T.; Li, T.-S.; Hamano, K. JAB1 Regulates Unphosphorylated STAT3 DNA-Binding Activity through Protein–Protein Interaction in Human Colon Cancer Cells. Biochem. Biophys. Res. Commun. 2013, 438, 513–518.
- Lobo, M.V.T.; Martín, M.E.; Pérez, M.I.; Alonso, F.J.M.; Redondo, C.; Álvarez, M.I.; Salinas, M. Levels, Phosphorylation Status and Cellular Localization of Translational Factor EIF2 in Gastrointestinal Carcinomas. Histochem. J. 2000, 32, 139–150.
- Tanabe, H.; Kuribayashi, K.; Tsuji, N.; Tanaka, M.; Kobayashi, D.; Watanabe, N. Sesamin Induces Autophagy in Colon Cancer Cells by Reducing Tyrosine Phosphorylation of EphA1 and EphB2. Int. J. Oncol. 2011, 39, 33–40.
- Park, H.-K.; Kim, I.-H.; Kim, J.; Nam, T.-J. Induction of Apoptosis and the Regulation of ErbB Signaling by Laminarin in HT-29 Human Colon Cancer Cells. Int. J. Mol. Med. 2013, 32, 291–295.
- Laferrière, J.; Houle, F.; Taher, M.M.; Valerie, K.; Huot, J. Transendothelial Migration of Colon Carcinoma Cells Requires Expression of E-Selectin by Endothelial Cells and Activation of Stress-Activated Protein Kinase-2 (SAPK2/P38) in the Tumor Cells. J. Biol. Chem. 2001, 276, 33762–33772.
- An, H.; Choi, E.; Kim, J.; Hong, S.-W.; Moon, J.-H.; Shin, J.-S.; Ha, S.-H.; Kim, K.P.; Hong, Y.; Lee, J.-L.; et al. INCB018424 Induces Apoptotic Cell Death through the Suppression of PJAK1 in Human Colon Cancer Cells. Neoplasma 2014, 61, 56–62.
- Tai, C.-J.; Lee, C.-H.; Chen, H.-C.; Wang, H.-K.; Jiang, M.-C.; Su, T.-C.; Shen, K.-H.; Lin, S.-H.; Yeh, C.-M.; Chen, C.-J.; et al. High Nuclear Expression of Phosphorylated Extracellular Signal–Regulated Kinase in Tumor Cells in Colorectal Glands Is Associated with Poor Outcome in Colorectal Cancer. Ann. Diagn. Pathol. 2013, 17, 165–171.
- Hua, H.; Chen, W.; Shen, L.; Sheng, Q.; Teng, L. Honokiol Augments the Anti-Cancer Effects of Oxaliplatin in Colon Cancer Cells. Acta Biochim. Biophys. Sin. (Shanghai) 2013, 45, 773–779.
- Wang, B.; Wang, W.; Niu, W.; Liu, E.; Liu, X.; Wang, J.; Peng, C.; Liu, S.; Xu, L.; Wang, L.; et al. SDF-1/CXCR4 Axis Promotes Directional Migration of Colorectal Cancer Cells through Upregulation of Integrin Avβ6. Carcinogenesis 2014, 35, 282–291.
- Wei, S.-C.; Tsao, P.-N.; Weng, M.-T.; Cao, Z.; Wong, J.-M. Flt-1 in Colorectal Cancer Cells Is Required for the Tumor Invasive Effect of Placental Growth Factor through a P38-MMP9 Pathway. J. Biomed. Sci. 2013, 20, 39.
- Lee, S.U.G.H.; Lee, J.W.O.O.; Soung, Y.H.W.A.; Kim, S.U.Y.; Nam, S.U.K.W.O.O.; Park, W.O.N.S.; Kim, S.H.O.; Yoo, N.A.M.J.I.N.; Lee, J.Y. Colorectal Tumors Frequently Express Phosphorylated Mitogen-Activated Protein Kinase. Apmis 2004, 112, 233–238.
- Wang, D.; Lao, W.-F.; Kuang, Y.-Y.; Geng, S.-M.; Mo, L.-J.; He, C. A Novel Variant of the RON Receptor Tyrosine Kinase Derived from Colorectal Carcinoma Cells Which Lacks Tyrosine Phosphorylation but Induces Cell Migration. Exp. Cell Res. 2012, 318, 2548–2558.
- Čačev, T.; Aralica, G.; Lončar, B.; Kapitanović, S. Loss of NF2/Merlin Expression in Advanced Sporadic Colorectal Cancer. Cell. Oncol. 2014, 37, 69–77.
- Josse, C.; Bouznad, N.; Geurts, P.; Irrthum, A.; Huynh-Thu, V.A.; Servais, L.; Hego, A.; Delvenne, P.; Bours, V.; Oury, C. Identification of a MicroRNA Landscape Targeting the PI3K/Akt Signaling Pathway in Inflammation-Induced Colorectal Carcinogenesis. Am. J. Physiol.-Gastrointest. Liver Physiol. 2013, 306, G229–G243.
- Jihane, B.; Dany, C.; Robert, H.; Isabelle, J.-E.; Marc, P. Ubiquitin-Independent Proteasomal Degradation of Fra-1 Is Antagonized by Erk1/2 Pathway-Mediated Phosphorylation of a Unique C-Terminal Destabilizer. Mol. Cell Biol. 2007, 27, 3936–3950.
- Huang, C.-C.; Wu, D.-W.; Lin, P.-L.; Lee, H. Paxillin Promotes Colorectal Tumor Invasion and Poor Patient Outcomes via ERK-Mediated Stabilization of Bcl-2 Protein by Phosphorylation at Serine 87. Oncotarget 2015, 6, 8698.
- Fang, L.; Lu, W.; Choi, H.H.; Yeung, S.-C.J.; Tung, J.-Y.; Hsiao, C.-D.; Fuentes-Mattei, E.; Menter, D.; Chen, C.; Wang, L.; et al. ERK2-Dependent Phosphorylation of CSN6 Is Critical in Colorectal Cancer Development. Cancer Cell 2015, 28, 183–197.
- Doller, A.; Winkler, C.; Azrilian, I.; Schulz, S.; Hartmann, S.; Pfeilschifter, J.; Eberhardt, W. High-Constitutive HuR Phosphorylation at Ser 318 by PKCδ Propagates Tumor Relevant Functions in Colon Carcinoma Cells. Carcinogenesis 2011, 32, 676–685.
- Hashimoto, Y.; Parsons, M.; Adams, J.C. Dual Actin-Bundling and Protein Kinase C-Binding Activities of Fascin Regulate Carcinoma Cell Migration Downstream of Rac and Contribute to Metastasis. Mol. Biol. Cell 2007, 18, 4591–4602.
- Lee, C.-C.; Lin, Y.-H.; Chang, W.-H.; Lin, P.-C.; Wu, Y.-C.; Chang, J.-G. Squamocin Modulates Histone H3 Phosphorylation Levels and Induces G1 Phase Arrest and Apoptosis in Cancer Cells. BMC Cancer 2011, 11, 58.
- Cabot, D.; Brun, S.; Paco, N.; Ginesta, M.M.; Gendrau-Sanclemente, N.; Abuasaker, B.; Ruiz-Fariña, T.; Barceló, C.; Cuatrecasas, M.; Bosch, M.; et al. KRAS Phosphorylation Regulates Cell Polarization and Tumorigenic Properties in Colorectal Cancer. Oncogene 2021, 40, 5730–5740.
- Huang, M.-J.; Wang, P.-N.; Huang, J.; Zhang, X.-W.; Wang, L.; Liu, H.; Wang, J.-P. Expression and Clinicopathological Significance of Serine257/Threonine261 Phosphorylated MKK4 in Colorectal Carcinoma. Zhonghua Yi Xue Za Zhi 2013, 93, 746–750.
- Hu, B.; Ren, D.; Su, D.; Lin, H.; Xian, Z.; Wan, X.; Zhang, J.; Fu, X.; Jiang, L.; Diao, D.; et al. Expression of the Phosphorylated MEK5 Protein Is Associated with TNM Staging of Colorectal Cancer. BMC Cancer 2012, 12, 127.
- Lewander, A.; Gao, J.; Carstensen, J.; Arbman, G.; Zhang, H.; Sun, X.-F. NF-ΚB P65 Phosphorylated at Serine-536 Is an Independent Prognostic Factor in Swedish Colorectal Cancer Patients. Int. J. Color. Dis. 2012, 27, 447–452.
- Jeong, J.B.; Yang, X.; Clark, R.; Choi, J.; Baek, S.J.; Lee, S.-H. A Mechanistic Study of the Proapoptotic Effect of Tolfenamic Acid: Involvement of NF-ΚB Activation. Carcinogenesis 2013, 34, 2350–2360.
- Li, Y.; Park, J.; Piao, L.; Kong, G.; Kim, Y.; Park, K.A.; Zhang, T.; Hong, J.; Hur, G.M.; Seok, J.H.; et al. PKB-Mediated PHF20 Phosphorylation on Ser291 Is Required for P53 Function in DNA Damage. Cell. Signal. 2013, 25, 74–84.
- Li, N.; Lorenzi, F.; Kalakouti, E.; Normatova, M.; Jadidi, R.; Tomlinson, I.; Nateri, A. FBXW7-Mutated Colorectal Cancer Cells Exhibit Aberrant Expression of Phosphorylated-P53 at Serine-15. Oncotarget 2015, 6, 9240.
- Lee, J.M.; Kim, I.S.; Kim, H.; Lee, J.S.; Kim, K.; Yim, H.Y.; Jeong, J.; Kim, J.H.; Kim, J.-Y.; Lee, H.; et al. RORα Attenuates Wnt/β-Catenin Signaling by PKCα-Dependent Phosphorylation in Colon Cancer. Mol. Cell 2010, 37, 183–195.
- Lee, Y.-H.; Kim, S.-J.; Fang, X.; Song, N.-Y.; Kim, D.-H.; Suh, J.; Na, H.-K.; Kim, K.-O.; Baek, J.-H.; Surh, Y.-J. JNK-Mediated Ser27 Phosphorylation and Stabilization of SIRT1 Promote Growth and Progression of Colon Cancer through Deacetylation-Dependent Activation of Snail. Mol. Oncol. 2022, 16, 1555–1571.
- Zhao, M.; Gjerset, R.A. Topoisomerase-I PS506 as a Dual Function Cancer Biomarker. PLoS ONE 2015, 10, e0134929.
- Simoneau, B.; Houle, F.; Huot, J. Regulation of Endothelial Permeability and Transendothelial Migration of Cancer Cells by Tropomyosin-1 Phosphorylation. Vasc. Cell 2012, 4, 18.
- Zykova, T.A.; Zhu, F.; Wang, L.; Li, H.; Bai, R.; Lim, D.Y.; Yao, K.; Bode, A.M.; Dong, Z. The T-LAK Cell-Originated Protein Kinase Signal Pathway Promotes Colorectal Cancer Metastasis. EBioMedicine 2017, 18, 73–82.
- Tomasi, M.L.; Tomasi, I.; Ramani, K.; Pascale, R.M.; Xu, J.; Giordano, P.; Mato, J.M.; Lu, S.C. S-Adenosyl Methionine Regulates Ubiquitin-Conjugating Enzyme 9 Protein Expression and Sumoylation in Murine Liver and Human Cancers. Hepatology 2012, 56, 982–993.
- Ohara, M.; Ohara, K.; Kumai, T.; Ohkuri, T.; Nagato, T.; Hirata-Nozaki, Y.; Kosaka, A.; Nagata, M.; Hayashi, R.; Harabuchi, S.; et al. Phosphorylated Vimentin as an Immunotherapeutic Target against Metastatic Colorectal Cancer. Cancer Immunol. Immunother. 2020, 69, 989–999.
- Li, J.; Hu, H.; Lang, Q.; Zhang, H.; Huang, Q.; Wu, Y.; Yu, L. A Thienopyrimidine Derivative Induces Growth Inhibition and Apoptosis in Human Cancer Cell Lines via Inhibiting Aurora B Kinase Activity. Eur. J. Med. Chem. 2013, 65, 151–157.
- Dey, H.; Liu, Z.-R. Phosphorylation of P68 RNA Helicase by P38 MAP Kinase Contributes to Colon Cancer Cells Apoptosis Induced by Oxaliplatin. BMC Cell Biol. 2012, 13, 27.
- Morris, J.F.; Sul, J.-Y.; Kim, M.-S.; Klein-Szanto, A.J.; Schochet, T.; Rustgi, A.; Eberwine, J.H. Elk-1 Phosphorylated at Threonine-417 Is Present in Diverse Cancers and Correlates with Differentiation Grade of Colonic Adenocarcinoma. Hum. Pathol. 2013, 44, 766–776.
- Pineda, C.T.; Ramanathan, S.; Fon Tacer, K.; Weon, J.L.; Potts, M.B.; Ou, Y.-H.; White, M.A.; Potts, P.R. Degradation of AMPK by a Cancer-Specific Ubiquitin Ligase. Cell 2015, 160, 715–728.
- Janoštiak, R.; Tolde, O.; Brůhová, Z.; Novotný , M.; Hanks, S.K.; Rösel, D.; Brábek, J. Tyrosine Phosphorylation within the SH3 Domain Regulates CAS Subcellular Localization, Cell Migration, and Invasiveness. Mol. Biol. Cell 2011, 22, 4256–4267.
- Zhang, P.; Guo, A.; Possemato, A.; Wang, C.; Beard, L.; Carlin, C.; Markowitz, S.D.; Polakiewicz, R.D.; Wang, Z. Identification and Functional Characterization of P130Cas as a Substrate of Protein Tyrosine Phosphatase Nonreceptor 14. Oncogene 2013, 32, 2087–2095.
- Joshi, B.; Strugnell, S.S.; Goetz, J.G.; Kojic, L.D.; Cox, M.E.; Griffith, O.L.; Chan, S.K.; Jones, S.J.; Leung, S.-P.; Masoudi, H.; et al. Phosphorylated Caveolin-1 Regulates Rho/ROCK-Dependent Focal Adhesion Dynamics and Tumor Cell Migration and Invasion. Cancer Res. 2008, 68, 8210–8220.
- Uchiyama, T.; Takahashi, H.; Sugiyama, M.; Sakai, E.; Endo, H.; Hosono, K.; Yoneda, K.; Yoneda, M.; Inamori, M.; Nagashima, Y.; et al. Leptin Receptor Is Involved in STAT3 Activation in Human Colorectal Adenoma. Cancer Sci. 2011, 102, 367–372.
- Xu, Y.; Jin, J.; Zhang, W.; Zhang, Z.; Gao, J.; Liu, Q.; Zhou, C.; Xu, Q.; Shi, H.; Hou, Y.; et al. EGFR/MDM2 Signaling Promotes NF-ΚB Activation via PPARγ Degradation. Carcinogenesis 2016, 37, 215–222.
- Cristóbal, I.; Manso, R.; Rincón, R.; Caramés, C.; Zazo, S.; del Pulgar, T.G.; Cebrián, A.; Madoz-Gúrpide, J.; Rojo, F.; García-Foncillas, J. Phosphorylated Protein Phosphatase 2A Determines Poor Outcome in Patients with Metastatic Colorectal Cancer. Br. J. Cancer 2014, 111, 756–762.
- Matkowskyj, K.A.; Keller, K.; Glover, S.; Kornberg, L.; Tran-Son-Tay, R.; Benya, R.V. Expression of GRP and Its Receptor in Well-Differentiated Colon Cancer Cells Correlates with the Presence of Focal Adhesion Kinase Phosphorylated at Tyrosines 397 and 407. J. Histochem. Cytochem. 2003, 51, 1041–1048.
- Golas, J.M.; Lucas, J.; Etienne, C.; Golas, J.; Discafani, C.; Sridharan, L.; Boghaert, E.; Arndt, K.; Ye, F.; Boschelli, D.H.; et al. SKI-606, a Src/Abl Inhibitor with In Vivo Activity in Colon Tumor Xenograft Models. Cancer Res. 2005, 65, 5358–5364.
- Fiordalisi, J.J.; Dewar, B.J.; Graves, L.M.; Madigan, J.P.; Cox, A.D. Src-Mediated Phosphorylation of the Tyrosine Phosphatase PRL-3 Is Required for PRL-3 Promotion of Rho Activation, Motility and Invasion. PLoS ONE 2013, 8, e64309.
- Zhao, Y.; Zhang, X.; Guda, K.; Lawrence, E.; Sun, Q.; Watanabe, T.; Iwakura, Y.; Asano, M.; Wei, L.; Yang, Z.; et al. Identification and Functional Characterization of Paxillin as a Target of Protein Tyrosine Phosphatase Receptor T. Proc. Natl. Acad. Sci. USA 2010, 107, 2592–2597.
- Serrels, A.; Macpherson, I.R.J.; Evans, T.R.J.; Lee, F.Y.; Clark, E.A.; Sansom, O.J.; Ashton, G.H.; Frame, M.C.; Brunton, V.G. Identification of Potential Biomarkers for Measuring Inhibition of Src Kinase Activity in Colon Cancer Cells Following Treatment with Dasatinib. Mol. Cancer 2006, 5, 3014–3022.
- Zhang, P.; Zhao, Y.; Zhu, X.; Sedwick, D.; Zhang, X.; Wang, Z. Cross-Talk between Phospho-STAT3 and PLCγ1 Plays a Critical Role in Colorectal Tumorigenesis. Mol. Cancer Res. 2011, 9, 1418–1428.
- Cai, Q.; Lin, J.; Wei, L.; Zhang, L.; Wang, L.; Zhan, Y.; Zeng, J.; Xu, W.; Shen, A.; Hong, Z.; et al. Hedyotis Diffusa Willd Inhibits Colorectal Cancer Growth in Vivo via Inhibition of STAT3 Signaling Pathway. Int. J. Mol. Sci. 2012, 13, 6117–6128.
- Hu, X.; Dutta, P.; Tsurumi, A.; Li, J.; Wang, J.; Land, H.; Li, W.X. Unphosphorylated STAT5A Stabilizes Heterochromatin and Suppresses Tumor Growth. Proc. Natl. Acad. Sci. USA 2013, 110, 10213–10218.
- Ward, P.S.; Thompson, C.B. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell 2012, 21, 297–308.
- Phan, L.M.; Yeung, S.-C.J.; Lee, M.-H. Cancer Metabolic Reprogramming: Importance, Main Features, and Potentials for Precise Targeted Anti-Cancer Therapies. Cancer Biol. Med. 2014, 11, 1.
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674.
- Kieran, M.W.; Kalluri, R.; Cho, Y.-J. The VEGF Pathway in Cancer and Disease: Responses, Resistance, and the Path Forward. Cold Spring Harb. Perspect. Med. 2012, 2, a006593.
- Shibuya, M. Vascular Endothelial Growth Factor and Its Receptor System: Physiological Functions in Angiogenesis and Pathological Roles in Various Diseases. J. Biochem. 2013, 153, 13–19.
More
Information
Subjects:
Genetics & Heredity
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Revisions:
3 times
(View History)
Update Date:
06 Feb 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




