
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Clint Gray | -- | 2208 | 2023-01-30 20:10:00 | | | |
| 2 | Jessie Wu | Meta information modification | 2208 | 2023-01-31 03:20:41 | | | | |
| 3 | Jessie Wu | Meta information modification | 2208 | 2023-01-31 03:22:38 | | | | |
| 4 | Jessie Wu | + 3 word(s) | 2211 | 2023-01-31 03:28:23 | | |
Video Upload Options
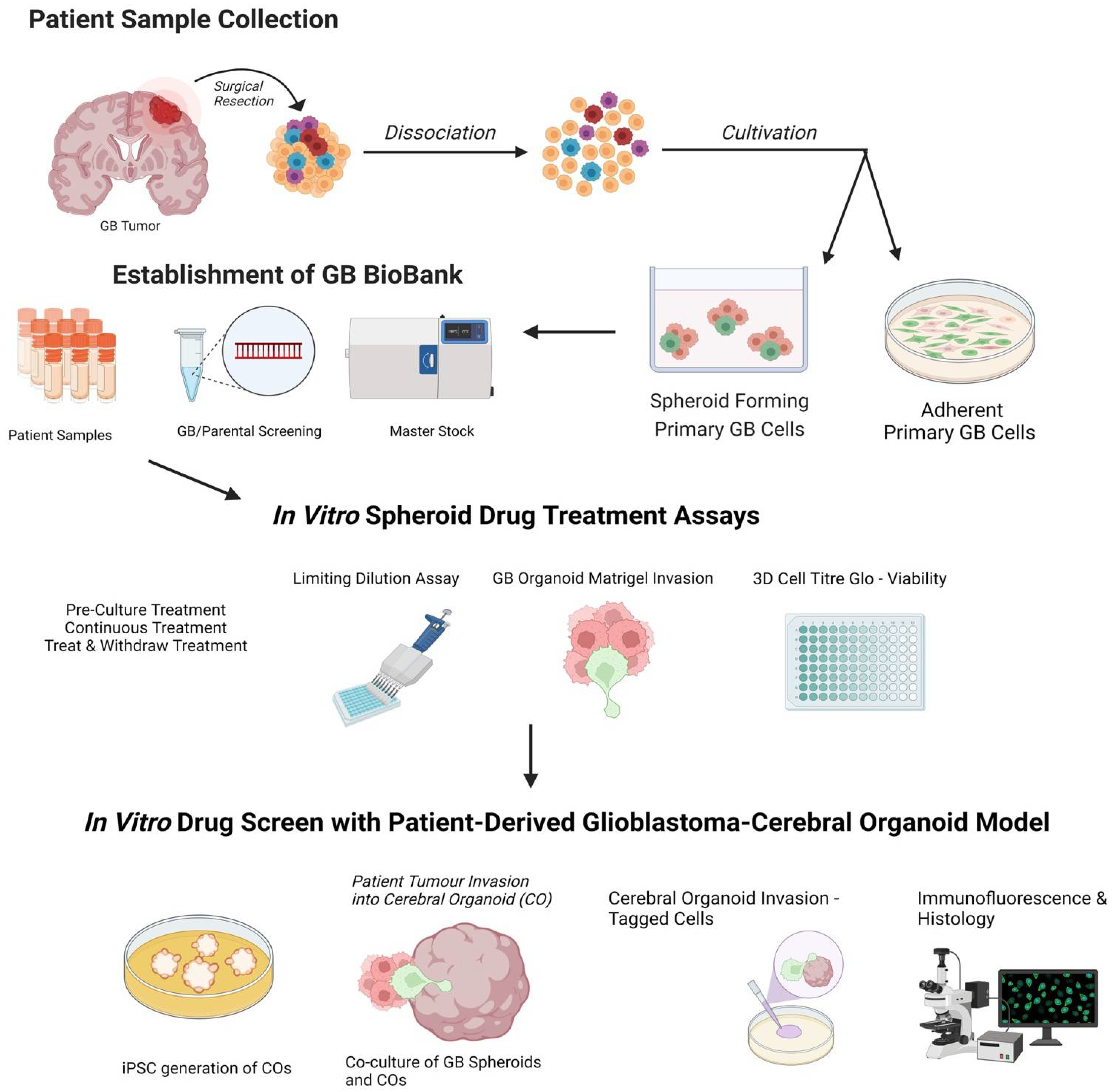
Glioblastoma, a grade IV astrocytoma, is regarded as the most aggressive primary brain tumour with an overall median survival of 16.0 months following the standard treatment regimen of surgical resection, followed by radiotherapy and chemotherapy with temozolomide. The ability to understand and manipulate complex cancers such as glioblastoma requires disease models to be clinically and translationally relevant and encompass the cellular heterogeneity of such cancers. Therefore, brain cancer research models need to aim to recapitulate glioblastoma stem cell function, whilst remaining amenable for analysis. The development of 3D cultures has overcome some of these challenges, and cerebral organoids are emerging as cutting-edge tools in glioblastoma research. The opportunity to generate cerebral organoids via induced pluripotent stem cells, and to perform co-cultures with patient-derived cancer stem cells (GLICO model), has enabled the analysis of cancer development in a context that better mimics brain tissue architecture.
1. Introduction
2. Patient-Derived Glioblastoma Organoids
3. Drug Screening Using a Patient-Derived Glioblastoma-Cerebral Organoid Model
References
- Huse, J.T.; Holland, E.; DeAngelis, L.M. Glioblastoma: Molecular Analysis and Clinical Implications. Annu. Rev. Med. 2013, 64, 59–70.
- Stupp, R.; Taillibert, S.; Kanner, A.; Read, W.; Steinberg, D.M.; Lhermitte, B.; Toms, S.; Idbaih, A.; Ahluwalia, M.S.; Fink, K.; et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma. JAMA 2017, 318, 2306–2316.
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. New Engl. J. Med. 2005, 352, 987–996.
- Cloughesy, T.F.; Cavenee, W.K.; Mischel, P.S. Glioblastoma: From Molecular Pathology to Targeted Treatment. Annu. Rev. Pathol. Mech. Dis. 2014, 9, 1–25.
- Linkous, A.; Fine, H.A. Generating Patient-Derived Gliomas within Cerebral Organoids. STAR Protoc. 2020, 1, 100008.
- Andreatta, F.; Beccaceci, G.; Fortuna, N.; Celotti, M.; De Felice, D.; Lorenzoni, M.; Foletto, V.; Genovesi, S.; Rubert, J.; Alaimo, A. The Organoid Era Permits the Development of New Applications to Study Glioblastoma. Cancers 2020, 12, 3303.
- Kim, S.-S.; Pirollo, K.F.; Chang, E.H. Isolation and Culturing of Glioma Cancer Stem Cells. Curr. Protoc. Cell Biol. 2015, 67, 23.10.1–23.10.10.
- Rajcevic, U.; Petersen, K.; Knol, J.C.; Loos, M.; Bougnaud, S.; Klychnikov, O.; Li, K.W.; Pham, T.V.; Wang, J.; Miletic, H.; et al. iTRAQ-based Proteomics Profiling Reveals Increased Metabolic Activity and Cellular Cross-talk in Angiogenic Compared with Invasive Glioblastoma Phenotype. Mol. Cell Proteom. 2009, 8, 2595–2612.
- Bradshaw, A.; Wickremsekera, A.; Tan, S.T.; Peng, L.; Davis, P.F.; Itinteang, T. Cancer Stem Cell Hierarchy in Glioblastoma Multiforme. Front. Surg. 2016, 3, 21.
- Pearson, J.R.D.; Regad, T. Targeting cellular pathways in glioblastoma multiforme. Signal. Transduct. Target. 2017, 2, 17040.
- Alves, A.L.V.; Gomes, I.N.F.; Carloni, A.C.; Rosa, M.N.; da Silva, L.S.; Evangelista, A.F.; Reis, R.M.; Silva, V.A.O. Role of glioblastoma stem cells in cancer therapeutic resistance: A perspective on antineoplastic agents from natural sources and chemical derivatives. Stem Cell Res. 2021, 12, 206.
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134.
- Mao, H.; Lebrun, D.G.; Yang, J.; Zhu, V.F.; Li, M. Deregulated signaling pathways in glioblastoma multiforme: Molecular mechanisms and therapeutic targets. Cancer Investig. 2012, 30, 48–56.
- Melissaridou, S.; Wiechec, E.; Magan, M.; Jain, M.V.; Chung, M.K.; Farnebo, L.; Roberg, K. The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell fea.atures in head and neck cancer. Cancer Cell Int. 2019, 19, 16.
- Gunti, S.; Hoke, A.T.K.; Vu, K.P.; London, N.R. Organoid and Spheroid Tumor Models: Techniques and Applications. Cancers 2021, 13, 874.
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo. Cancer Res. 2016, 76, 2465–2477.
- Heddleston, J.M.; Li, Z.; McLendon, R.E.; Hjelmeland, A.B.; Rich, J.N. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle 2009, 8, 3274–3284.
- Quereda, V.; Hou, S.; Madoux, F.; Scampavia, L.; Spicer, T.P.; Duckett, D. A Cytotoxic Three-Dimensional-Spheroid, High-Throughput Assay Using Patient-Derived Glioma Stem Cells. Slas Discov. Adv. Sci. Drug Discov. 2018, 23, 842–849.
- Linkous, A.; Balamatsias, D.; Snuderl, M.; Edwards, L.; Miyaguchi, K.; Milner, T.; Reich, B.; Cohen-Gould, L.; Storaska, A.; Nakayama, Y.; et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep. 2019, 26, 3203–3211.e5.
- Auffinger, B.; Spencer, D.; Pytel, P.; Ahmed, A.U.; Lesniak, M.S. The role of glioma stem cells in chemotherapy resistance and glioblastoma multiforme recurrence. Expert Rev. Neurother. 2015, 15, 741–752.
- Ravi, V.M.; Will, P.; Kueckelhaus, J.; Sun, N.; Joseph, K.; Salié, H.; Vollmer, L.; Kuliesiute, U.; von Ehr, J.; Benotmane, J.K.; et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 2022, 40, 639–655.e13.
- Kilmister, E.J.; Tan, S.T. The Role of the Renin–Angiotensin System in the Cancer Stem Cell Niche. J Histochem. Cytochem. 2021, 69, 835–847.
- Geribaldi-Doldán, N.; Fernández-Ponce, C.; Quiroz, R.N.; Sánchez-Gomar, I.; Escorcia, L.G.; Velásquez, E.P.; Quiroz, E.N. The Role of Microglia in Glioblastoma. Front. Oncol. 2021, 10, 603495.
- Vinel, C.; Rosser, G.; Guglielmi, L.; Constantinou, M.; Pomella, N.; Zhang, X.; Boot, J.R.; Jones, T.A.; Millner, T.O.; Dumas, A.A.; et al. Comparative epigenetic analysis of tumour initiat.ting cells and syngeneic EPSC-derived neural stem cells in glioblastoma. Nat. Commun. 2021, 12, 6130.
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403.
- Azzarelli, R. Organoid Models of Glioblastoma to Study Brain Tumor Stem Cells. Front. Cell Dev. Biol. 2020, 8, 220.
- Ballav, S.; Deshmukh, A.J.; Siddiqui, S.; Aich, J.; Basu, S.; Ballav, S.; Deshmukh, A.J.; Siddiqui, S.; Aich, J.; Basu, S. Two-Dimensional and Three-Dimensional Cell Culture and Their Applications; IntechOpen: London, UK, 2021; ISBN 978-1-83969-446-2.
- Paolillo, M.; Comincini, S.; Schinelli, S. In Vitro Glioblastoma Models: A Journey into the Third Dimension. Cancers 2021, 13, 2449.
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218.
- Pollard, S.M.; Yoshikawa, K.; Clarke, I.D.; Danovi, D.; Stricker, S.; Russell, R.; Bayani, J.; Head, R.; Lee, M.; Bernstein, M.; et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 2009, 4, 568–580.
- Białkowska, K.; Komorowski, P.; Bryszewska, M.; Miłowska, K. Spheroids as a Type of Three-Dimensional Cell Cultures—Examples of Methods of Preparation and the Most Important Application. Int. J. Mol. Sci. 2020, 21, 6225.
- Azari, H.; Millette, S.; Ansari, S.; Rahman, M.; Deleyrolle, L.P.; Reynolds, B.A. Isolation and expansion of human glioblastoma multiforme tumor cells using the neurosphere assay. J. Vis. Exp. 2011, 30, e3633.
- Ramani, A.; Müller, L.; Ostermann, P.N.; Gabriel, E.; Abida-Islam, P.; Müller-Schiffmann, A.; Mariappan, A.; Goureau, O.; Gruell, H.; Walker, A.; et al. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020, 39, e106230.
- Yi, H.-G.; Jeong, Y.H.; Kim, Y.; Choi, Y.-J.; Moon, H.E.; Park, S.H.; Kang, K.S.; Bae, M.; Jang, J.; Youn, H.; et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 2019, 3, 509–519.
- Ogawa, J.; Pao, G.M.; Shokhirev, M.N.; Verma, I.M. Glioblastoma Model Using Human Cerebral Organoids. Cell Rep. 2018, 23, 1220–1229.
- Bian, S.; Repic, M.; Guo, Z.; Kavirayani, A.; Burkard, T.; Bagley, J.A.; Krauditsch, C.; Knoblich, J.A. Genetically engineered cerebral organoids model brain tumour formation. Nat. Methods 2018, 15, 631–639.
- Baskar, G.; Palaniyandi, T.; Viswanathan, S.; Rajendran, B.K.; Ravi, M.; Sivaji, A. Development of patient derived organoids for cancer drug screening applications. Acta Histochem. 2022, 124, 151895.
- Da Silva, B.; Mathew, R.K.; Polson, E.S.; Williams, J.; Wurdak, H. Spontaneous Glioblastoma Spheroid Infiltration of Early-Stage Cerebral Organoids Models Brain Tumor Invasion. SLAS Discov. 2018, 23, 862–868.
- Hicks, W.H.; Bird, C.E.; Gattie, L.C.; Shami, M.E.; Traylor, J.I.; Shi, D.D.; McBrayer, S.K.; Abdullah, K.G. Creation and Development of Patient-Derived Organoids for Therapeutic Screening in Solid Cancer. Curr. Stem Cell Rep. 2022, 8, 107–117.
- Silvia, N.; Dai, G. Cerebral organoids as a model for glioblastoma multiforme. Curr. Opin. Biomed. Eng. 2020, 13, 152–159.
- Mariappan, A.; Goranci-Buzhala, G.; Ricci-Vitiani, L.; Pallini, R.; Gopalakrishnan, J. Trends and challenges in modeling glioma using 3D human brain organoids. Cell Death Differ. 2021, 28, 15–23.
- Goranci-Buzhala, G.; Mariappan, A.; Gabriel, E.; Ramani, A.; Ricci-Vitiani, L.; Buccarelli, M.; D’Alessandris, Q.G.; Pallini, R.; Gopalakrishnan, J. Rapid and Efficient Invasion Assay of Glioblastoma in Human Brain Organoids. Cell Rep. 2020, 31, 107738.
- Andrews, M.G.; Kriegstein, A.R. Challenges of Organoid Research. Annu. Rev. Neurosci. 2022, 45, 23–39.
- Zhang, W.; Jiang, J.; Xu, Z.; Yan, H.; Tang, B.; Liu, C.; Chen, C.; Meng, Q. Microglia-containing human brain organoids for the study of brain development and pathology. Mol. Psychiatry 2022.
- Cakir, B.; Xiang, Y.; Tanaka, Y.; Kural, M.H.; Parent, M.; Kang, Y.-J.; Chapeton, K.; Patterson, B.; Yuan, Y.; He, C.-S.; et al. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods 2019, 16, 1169–1175.
- Ormel, P.R.; Vieira de Sá, R.; van Bodegraven, E.J.; Karst, H.; Harschnitz, O.; Sneeboer, M.A.M.; Johansen, L.E.; van Dijk, R.E.; Scheefhals, N.; Berdenis van Berlekom, A.; et al. Microglia innately develop within cerebral organoids. Nat. Commun. 2018, 9, 4167.
- Pham, M.T.; Pollock, K.M.; Rose, M.D.; Cary, W.A.; Stewart, H.R.; Zhou, P.; Nolta, J.A.; Waldau, B. Generation of human vascularized brain organoids. Neuroreport 2018, 29, 588–593.
- Rybin, M.J.; Ivan, M.E.; Ayad, N.G.; Zeier, Z. Organoid Models of Glioblastoma and Their Role in Drug Discovery. Front. Cell. Neurosci. 2021, 15, 605255.
- Coombe, D.R.; Nakhoul, A.M.; Stevenson, S.M.; Peroni, S.E.; Sanderson, C.J. Expressed luciferase viability assay (ELVA) for the measurement of cell growth and viability. J. Immunol. Methods 1998, 215, 145–150.




