
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Dilip Ghosh | -- | 3132 | 2023-01-16 17:56:51 | | | |
| 2 | Camila Xu | Meta information modification | 3132 | 2023-01-17 02:51:32 | | |
Video Upload Options
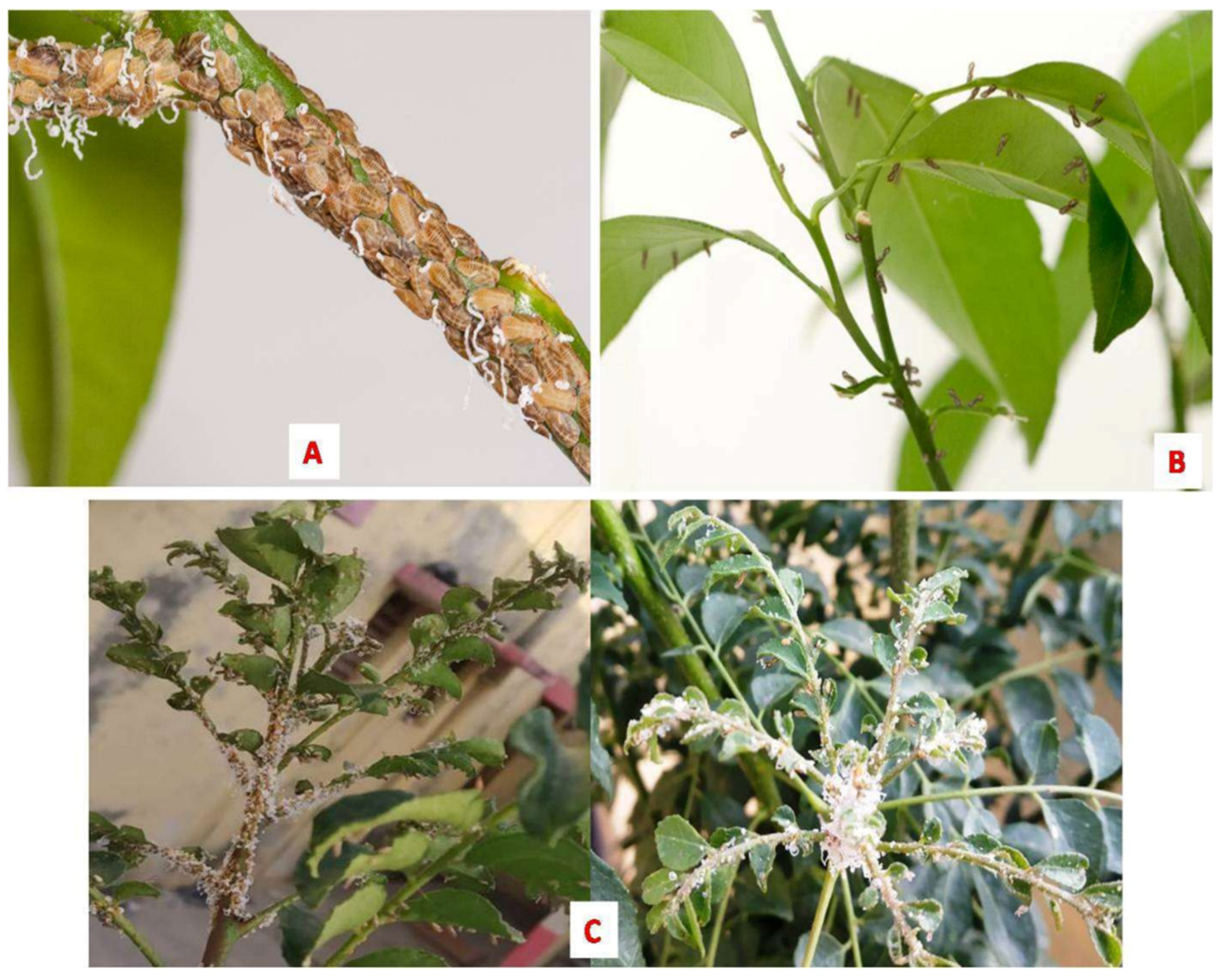
Huanglongbing (HLB, aka citrus greening), one of the most devastating diseases of citrus, has wreaked havoc on the global citrus industry in recent decades. The culprit behind such a gloomy scenario is the phloem-limited bacteria “Candidatus Liberibacter asiaticus” (CLas), which are transmitted via psyllid. To date, there are no effective long-termcommercialized control measures for HLB, making it increasingly difficult to prevent the disease spread. To combat HLB effectively, introduction of multipronged management strategies towards controlling CLas population within the phloem system is deemed necessary. This entry presents a comprehensive review of up-to-date scientific information about HLB, including currently available management practices and unprecedented challenges associated with the disease control. Additionally, a triangular disease management approach has been introduced targeting pathogen, host, and vector. Pathogen-targeting approaches include (i) inhibition of important proteins of CLas, (ii) use of the most efficient antimicrobial or immunity-inducing compounds to suppress the growth of CLas, and (iii) use of tools to suppress or kill the CLas. Approaches for targeting the host include (i) improvement of the host immune system, (ii) effective use of transgenic variety to build the host’s resistance against CLas, and (iii) induction of systemic acquired resistance. Strategies for targeting the vector include (i) chemical and biological control and (ii) eradication of HLB-affected trees. Finally, a hypothetical model for integrated disease management has been discussed to mitigate the HLB pandemic.
1. Introduction
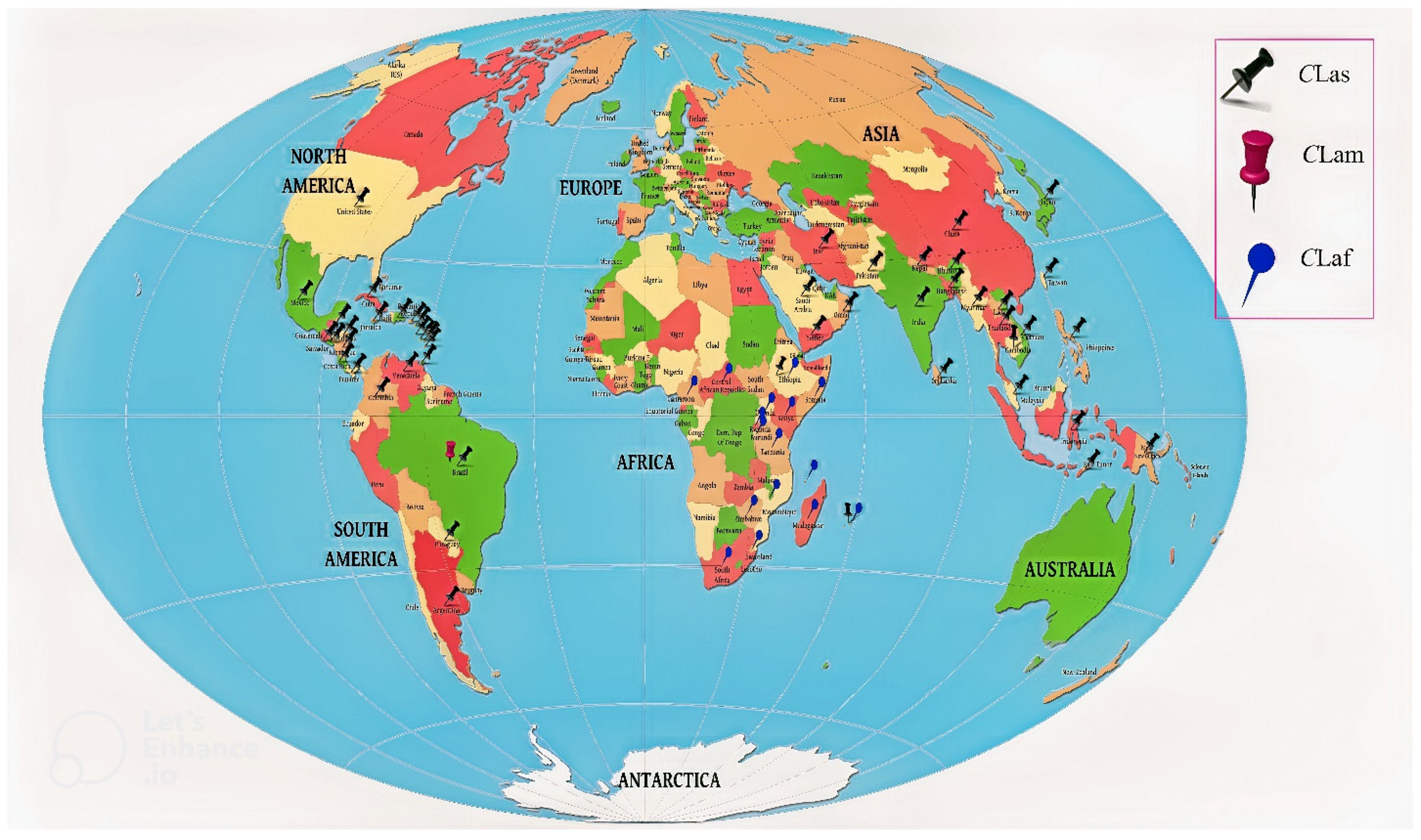
| Sr. No | Continent/Country/Region | Distribution of HLB | Causal Organism | Reference |
|---|---|---|---|---|
| Asia | ||||
| 1 | Bangladesh | Present | Ca. Liberibacter asiaticus | [21] |
| 2 | Bhutan | Present | Ca. Liberibacter asiaticus | [22] |
| 3 | Cambodia | Present | Ca. Liberibacter asiaticus | [23] |
| 4 | China | Present | Ca. Liberibacter asiaticus | [22] |
| 5 | India | Present, Widespread | Ca. Liberibacter asiaticus | [24] |
| 6 | Indonesia | Present | Ca. Liberibacter asiaticus | [25] |
| 7 | Iran | Present, Localized | Ca. Liberibacter asiaticus | [26] |
| 8 | Japan | Present | Ca. Liberibacter asiaticus | [27] |
| 9 | Laos | Present | Ca. Liberibacter asiaticus | [23] |
| 10 | Malaysia | Present, Localized | Ca. Liberibacter asiaticus | [24] |
| 11 | Myanmar | Present | Ca. Liberibacter asiaticus | [23] |
| 12 | Nepal | Present, Widespread | Ca. Liberibacter asiaticus | [28] |
| 13 | Oman | Present, Localized | Ca. Liberibacter asiaticus | [22] |
| 14 | Pakistan | Present | Ca. Liberibacter asiaticus | [29] |
| 15 | Philippines | Present, Widespread | Ca. Liberibacter asiaticus | [29] |
| 16 | Saudi Arabia | Present, Localized | Ca. Liberibacter asiaticus | [30] |
| 17 | Sri Lanka | Present | Ca. Liberibacter asiaticus | [22] |
| 18 | Taiwan | Present, Widespread | Ca. Liberibacter asiaticus | [22] |
| 19 | Thailand | Present | Ca. Liberibacter asiaticus | [31] |
| 20 | Vietnam | Present, Localized | Ca. Liberibacter asiaticus | [32] |
| 21 | Yemen | Present, Localized | Ca. Liberibacter asiaticus | [30] |
| North America | Ca. Liberibacter asiaticus | |||
| 22 | Barbados | Present, Localized | Ca. Liberibacter asiaticus | [22] |
| 23 | Belize | Present, Localized | Ca. Liberibacter asiaticus | [17] |
| 24 | Costa Rica | Present, Localized | Ca. Liberibacter asiaticus | [22] |
| 25 | Cuba | Present, Widespread | Ca. Liberibacter asiaticus | [22] |
| 26 | Dominica | Present, Few occurrences | Ca. Liberibacter asiaticus | [22] |
| 27 | Dominican Republic | Present, Localized | Ca. Liberibacter asiaticus | [22] |
| 28 | El Salvador | Present | Unknown | [33] |
| 29 | Guadeloupe | Present, Localized | Unknown | [34] |
| 30 | Guatemala | Present | Ca. Liberibacter asiaticus | [22] |
| 31 | Honduras | Present | Ca. Liberibacter asiaticus | [22] |
| 32 | Jamaica | Present, Widespread | Ca. Liberibacter asiaticus | [22] |
| 33 | Martinique | Present, Localized | Ca. Liberibacter asiaticus | [34] |
| 34 | Mexico | Present, Localized | Ca. Liberibacter asiaticus | [22] |
| 35 | Nicaragua | Present | Ca. Liberibacter asiaticus | [22] |
| 36 | Panama | Present, Localized | Ca. Liberibacter asiaticus | [22] |
| 37 | Puerto Rico | Present | Ca. Liberibacter asiaticus | [22] |
| 38 | Trinidad and Tobago | Present, Localized | Ca. Liberibacter asiaticus | [22] |
| 39 | U.S. Virgin Islands | Present, Few occurrences | Ca. Liberibacter asiaticus | [22] |
| 40 | United States | Present, Localized | Ca. Liberibacter asiaticus | [22] |
| South America | ||||
| 41 | Argentina | Present, Localized | Ca. Liberibacter asiaticus | [22] |
| 42 | Brazil | Present, Localized | Ca. Liberibacter americanus and Ca. Liberibacter asiaticus | [9] |
| 43 | Colombia | Present, Few occurrences | Ca.Liberibacter asiaticus | [22] |
| 44 | Paraguay | Present, Localized | Ca. Liberibacter asiaticus | [35] |
| 45 | Venezuela | Present | Ca. Liberibacter asiaticus | [36] |
| Africa | ||||
| 46 | Burundi | Present | Ca. Liberibacter africanus | [37] |
| 47 | Cameroon | Present | Ca. Liberibacter africanus | [37] |
| 48 | Central African Republic | Present | Ca. Liberibacter africanus | [37] |
| 49 | Comoros | Present | Ca. Liberibacter africanus | [22] |
| 50 | Eswatini | Present | Ca. Liberibacter africanus | [38] |
| 51 | Ethiopia | Present | Ca. Liberibacter africanus and Ca. Liberibacter asiaticus |
[37] |
| 52 | Kenya | Present | Ca. Liberibacter africanus | [29] |
| 53 | Madagascar | Present | Ca. Liberibacter africanus | [38] |
| 54 | Malawi | Present | Ca. Liberibacter africanus | [37] |
| 55 | Mauritius | Present | Ca. Liberibacter africanus and Ca. Liberibacter asiaticus |
[22] |
| 56 | Rwanda | Present | Ca. Liberibacter africanus | [37] |
| 57 | Somalia | Present | Ca. Liberibacter africanus | [22] |
| 58 | South Africa | Present, Localized | Ca. Liberibacter africanus | [39] |
| 59 | Tanzania | Present, Localized | Ca. Liberibacter africanus | [38] |
| 60 | Uganda | Present | Ca. Liberibacter africanus | [40] |
| 61 | Zimbabwe | Present, Localized | Ca. Liberibacter africanus | [29] |

2. Causative Agent, Genomics, and Pathogenesis Mechanism
| Sr. No | Candidatus Liberibacter spp. | Strain | Host | Sample Origin | Genome Size (Mb) | Number CDS Present in the Genome | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Candidatus Liberibacter asiaticus | A4 (CP010804.1) |
Citrus reticulata | China: Guangdong | 1.23025 | 1067 | [59] |
| 2 | Candidatus Liberibacter asiaticus | Gxpsy (CP004005.1) |
Diaphorinacitri | China: Guangxi | 1.26824 | 1094 | [57] |
| 3 | Candidatus Liberibacter asiaticus | JRPAMB1 (CP040636.1) |
Diaphorinacitri | USA: Florida | 1.23716 | 1072 | [62] |
| 4 | Candidatus Liberibacter asiaticus | TaiYZ2 (CP041385.1) | Citrus maxima | Thailand: Songkhla | 1.23062 | 1067 | [77] |
| 5 | Candidatus Liberibacter asiaticus | psy62 (CP001677.5) |
- | USA: Florida | 1.22732 | 1049 | [56] |
| 6 | Candidatus Liberibacter asiaticus | JXGC (CP019958.1) |
Citrus | China: Jiangxi | 1.22516 | 1033 | [60] |
| 7 | Candidatus Liberibacter asiaticus | Ishi-1 (AP014595.1) |
Diaphorinacitri | Japan: Ishigaki | 1.19085 | 1001 | [58] |
| 8 | Candidatus Liberibacter asiaticus | AHCA1 (CP029348.1) |
Diaphorinacitri | USA: California | 1.23375 | 1056 | [61] |
| 9 | Candidatus Liberibacter asiaticus | FL17 (JWHA00000000.1) |
Citrus | USA: Florida | 1.22725 | 1019 | [78] |
| 10 | Candidatus Liberibacter asiaticus | YNJS7C (QXDO00000000) |
Citrus | China: Yunnan | 1.25898 | 1102 | [79] |
| 11 | Candidatus Liberibacter asiaticus | YCPsy LIIM00000000) |
Diaphorinacitri | China: Guangdong | 1.233647 | 1037 | [80] |
| 12 | Candidatus Liberibacter asiaticus | LBR19TX2 (VTMA00000000 |
- | USA: Texas | 1.20275 | 1008 | [67] |
| 13 | Candidatus Liberibacter asiaticus | LBR23TX5 (VTMB00000000) |
- | USA: Texas | 1.20347 | 1009 | [67] |
| 14 | Candidatus Liberibacter asiaticus | AHCA17 (VNFL00000000) |
Citrus maxima | USA: California | 1.20862 | 1036 | [81] |
| 15 | Candidatus Liberibacter asiaticus | YNXP-1 (VIGA00000000) |
Cuscuta | China: Yunnan | 1.20707 | 1031 | - |
| 16 | Candidatus Liberibacter asiaticus | SGCA16 (VTLZ00000000) |
- | USA: San Gabriel | 1.20994 | 1015 | [67] |
| 17 | Candidatus Liberibacter asiaticus | JXGZ-1 (VIQL00000000) |
Cuscuta | China: JiangXi | 1.21799 | 1040 | - |
| 18 | Candidatus Liberibacter asiaticus | DUR1TX1 VTLT00000000.1) |
- | USA: Texas | 1.20629 | 1011 | [67] |
| 19 | Candidatus Liberibacter asiaticus | Mex8 (VTLU00000000.1) |
- | Mexico: Mexicali | 1.24313 | 1042 | [67] |
| 20 | Candidatus Liberibacter asiaticus | SGCA5 (LMTO00000000.1) |
Orange citrus | USA: San Gabriel | 1.20138 | 1001 | [80] |
| 21 | Candidatus Liberibacter asiaticus | CHUC (VTLV00000000) |
- | China | 1.20845 | 1032 | [67] |
| 22 | Candidatus Liberibacter asiaticus | TX2351 (MTIM00000000) |
Asian citrus psyllid | USA: Texas | 1.252 | 1129 | [82] |
| 23 | Candidatus Liberibacter asiaticus | GFR3TX3 (VTLR00000000) |
- | USA: Texas | 1.20932 | 1013 | [67] |
| 24 | Candidatus Liberibacter asiaticus | HHCA16 (VTLY00000000) |
- | USA: Hacienda Heights | 1.20705 | 1012 | [67] |
| 25 | Candidatus Liberibacter asiaticus | MFL16 (VTLX00000000) |
- | USA: Florida | 1.19922 | 1012 | [67] |
| 26 | Candidatus Liberibacter asiaticus | DUR2TX1 (VTLS00000000) |
- | USA: Texas | 1.21232 | 1009 | [67] |
| 27 | Candidatus Liberibacter asiaticus | CRCFL16 (VTLW00000000) |
- | USA: Florida | 1.20828 | 1028 | [67] |
| 28 | Candidatus Liberibacter asiaticus | HHCA (JMIL00000000.2) |
Citrus sp. | USA: Hacienda Heights | 1.15062 | 867 | [59] |
| 29 | Candidatus Liberibacter asiaticus | TX1712 (QEWL00000000) |
Citrus sinensis | USA: Texas | 1.20333 | 0 | [83] |
| 30 | Candidatus Liberibacter asiaticus | SGpsy (QFZJ00000000.1) |
Diaphorinacitri | USA: San Gabriel | 0.769888 | 0 | [61] |
| 31 | Candidatus Liberibacter asiaticus | SGCA1 (QFZT00000000.1) |
USA: San Gabriel | 0.233414 | 557 | [61] | |
| 32 | Candidatus Liberibacter asiaticus | YCPsy (LIIM00000000) |
Diaphorinacitri | Guangdong, China | 1.233647 | - | [80] |
| 33 | Candidatus Liberibacter asiaticus | PA19 (WOXD01000000) | Kinnow mandarin | Pakistan | 1.224156 | 1059 | [84] |
| 34 | Candidatus Liberibacter asiaticus | PA20 (WOUN01000000) | Kinnow mandarin | Pakistan | 1.226225 | 1062 | [84] |
| 35 | Candidatus Liberibacter asiaticus | CoFLP1 (CP054558.1) |
Diaphorinacitri | Colombia: Municipio Dibulla |
1.231639 | 1048 | [64] |
| 36 | Candidatus Liberibacter asiaticus | 9PA (JABDRZ000000000.1) | Citrus sinensis | Brazil (South America) | 1.231881 | - | [85] |
| 37 | Candidatus Liberibacter asiaticus | MFL16 (VTLX00000000) |
Citrus | USA: Florida | 1,199,225 bp | - | [67] |
| 38 | Candidatus Liberibacter asiaticus | CRCFL16 (VTLW00000000) | Citrus | USA: Florida | 1,208,280 bp | - | [67] |
| 39 | Candidatus Liberibacter asiaticus | ReuSP1 (CP061535.1) |
Diaphorinacitri | France: La Reunion | 1.230064 | 1043 | [65] |
| 40 | Candidatus Liberibacter asiaticus | Tabriz.3(JAKQYA000000000.1) | Elaeagnus angustifolia | Iran: East Azerbaijan, Tabriz | 1.22409 | 589 | - |
| 41 | Candidatus Liberibacter asiaticus | YNHK-2 (WUUB01000000.1) |
Citrus | China: Yunnan | 1.08957 | - | [86] |
| 42 | Candidatus Liberibacter asiaticus | A-SBCA19 (JADBIB010000000.1) |
Diaphorinacitri | USA: California, San Bernardino County | 1.18688 | 1067 | [87] |
| 43 | Candidatus Liberibacter americanus | Sao Paulo (NC_022793) | Citrus sinensis | Brazil | 1.1952 | 945 | [73] |
| 44 | Candidatus Liberibacter americanus | PW_SP | Catharanthus roseus | Brazil: Sao Paulo | 1.17607 | 924 | - |
| 45 | Candidatus Liberibacter africanus | PTSAPSY | Psyllid | South Africa: Pretoria | 1.19223 | 1036 | - |
Pathogen Virulence Factors
References
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014, 32, 656–662.
- Bové, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37.
- Gottwald, T.R. Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 2010, 48, 119–139.
- Reinking, O.A. Diseases of economic plants in Southern China. Philipp. Agric. 1919, 8, 109–135.
- Husain, M.A.; Nath, D. The citrus psylla (Diaphorinacitri, Kuw.) . Mem. Dept. Agric. India Entomol. Ser. 1927, 10, 5–27.
- Capoor, S.P. Decline of citrus in India. Bull. Natl. Inst. Sci. 1963, 24, 48–64.
- Beattie, G.A.C.; Holford, P.; Mabberley, D.J.; Haigh, A.M.; Bayer, R. Aspects and insights of Australia-Asia collaborative research on huanglongbing. In Proceedings of the International Workshop for the Prevention of Citrus Greening Disease in Severely Infected Areas, Ishigaki, Japan, 6–7 December 2006; Japanese Ministry of Agriculture, Forestry and Fisheries: Tokyo, Japan, 2006; pp. 47–64.
- Merwe, A.J.; Anderssen, F.G. Chromium and manganese toxicity. Is it important in Transvaal citrus greening. Farming S. Afr. 1937, 12, 439–440.
- Coletta-Filho, H.D.; Targon, M.; Takita, M.A.; De Negri, J.D.; Pompeu, J.; Machado, M.A.; do Amaral, A.M.; Muller, G.W. First report of the causal agent of Huanglongbing (Candidatus Liberibacter asiaticus) in Brazil. Plant Dis. 2004, 88, 1382.
- Halbert, S.E. The discovery of huanglongbing in Florida. In Proceedings of the Second International Citrus Canker and Huanglongbing Research Workshop, Orlando, FL, USA, 7–11 November 2005; p. H-3.
- Molki, B.; Call, D.R.; Ha, P.T.; Omsland, A.; Gang, D.R.; Lindemann, S.R.; Killiny, N.; Beyenal, H. Growth of ‘Candidatus Liberibacter asiaticus’ in a host-free microbial culture is associated with microbial community composition. Enzym. Microb. Technol. 2020, 142, 109691.
- da Graca, J.V.; Kunta, M.; Setamou, M.; Rascoe, J.; Li, W.; Nakhla, M.K.; Salas, B.; Bartels, D.W. Huanglongbing in Texas: Report on the first detections in commercial citrus. J. Citrus Pathol. 2015, 2, 27939.
- Kumagai, L.B.; LeVesque, C.S.; Blomquist, C.L.; Madishetty, K.; Guo, Y.; Woods, P.W.; Rooney-Latham, S.; Rascoe, J.; Gallindo, T.; Schnabel, D.; et al. First report of Candidatus Liberibacter asiaticus associated with citrus huanglongbing in California. Plant Dis. 2013, 97, 283.
- Halbert, S.E.; Manjunath, K.L.; Ramadugu, C.; Brodie, M.W.; Webb, S.E.; Lee, R.F. Trailers transporting organges to processing plants move asian citrus psyllids. Florida Entomol. 2010, 93, 33–38.
- Luis, M.; Collazo, C.; Llauger, R.; Blanco, E.; Pena, I.; López, D.; González, C.; Casín, J.C.; Batista, L.; Kitajima, E.; et al. Occurrence of citrus Huanglongbing in Cuba and association of the disease with Candidatus Liberibacter asciaticus. J. Plant Pathol. 2009, 91, 709–712.
- Oberheim, A.P.; Brown, S.E.; McLaughlin, W.A. The identification and distribution of citrus greening disease in Jamaica. In Proceedings of the 2nd International Research Conference on Huanglongbing, Orlando, FL, USA, 10–14 January 2011; p. 114.
- Manjunath, K.L.; Ramadugu, C.; Majil, V.M.; Williams, S.; Irey, M.; Lee, R.F. First report of the citrus huanglongbing associated bacterium ‘Candidatus Liberibacter asiaticus’ from sweet orange, mexican lime, and asian citrus psyllid in Belize. Plant Dis. 2010, 94, 781.
- Trujillo-Arriaga, J.; Sanchez, A.H.; Robles, G.P.; de la Rosa, A.A.; Delgadillo, V.I.; Márquez, S.M. Antecedantes y situacion actual de Huanglongbing de loscıtricosen Mexico. In Proceedings of the 1er Simposio Nacional Sobre Investigacion para el Manejo del Psılido Asiatico de losCıtricos y el HLB en Mexico, Monterrey, Mexico, 8–9 December 2010; pp. 1–7.
- Faghihi, M.M.; Salehi, M.; Bagheri, A.; Izadpanah, K. First report of citrus huanglongbing disease on orange in Iran. Plant Pathol. 2009, 58, 793.
- CABI. Citrus Huanglongbing (Greening) Disease (Citrus Greening). In Invasive Species Compendium; CABI International: Wallingford, UK, 2020.
- Catling, H.D.; Garnier, M.; Bove, J.M. Presence of citrus greening disease in Bangladesh and a new method for rapid diagnosis. FAO Plant Prot. Bull. 1978, 26, 16–18.
- EPPO (European and Mediterranean Plant Protection Organization). ‘Candidatus Liberibacter asiaticus’. EPPO Datasheets on Pests Recommended for Regulation. 2022. Available online: https://gd.eppo.int (accessed on 25 November 2022).
- Garnier, M.; Bové, J.M. Huanglongbing in Cambodia, Laos and Myanmar. In Proceedings of the 14th Conference of the International Organization of Citrus Virologists, Riverside, CA, USA, 2000; da Graça, J.V., Lee, R.F., Yokomi, R.K., Eds.; IOCV: Concordia, Argentina, 2000; Volume 14, pp. 378–380.
- Bové, J.M.; Garnier, M.; Ahlawat, Y.S.; Chakraborty, N.K.; Varma, A. Detection of the Asian strains of the greening BLO by DNA-DNA hybridization in Indian orchard trees and Malaysian Diaphorina Citri Psyllids. Int. Organ. Citrus Virol. Conf. Proc. 1993, 12, 1957–2010.
- Aubert, B.; Garnier, M.; Guillaumin, D.; Herbagyandono, B.; Setiobudi, L.; Nurhadi, F. Greening, a serious threat for the citrus productions of the Indonesian archipelago. Future prospects of integrated control. Fruits 1985, 40, 549–563.
- Salehi, M.; Faghihi, M.M.; Khanchezar, A.; Bagheree, A.; Izadpanah, K. Distributioin of citrus Huanglongbing disease and its vector in southern Iran. Iranian J. Plant Pathol. 2012, 48, 195–208.
- Miyakawa, T.; Tsuno, K. Occurrence of citrus greening disease in the southern islands of Japan. Jpn. J. Phytopathol. 1989, 55, 667–670.
- Regmi, C.; Garnier, M.; Bové, J.M. Detection of the Asian Huanglongbing (Greening) Liberobacter in Nepal by DNA-DNA hybridization. Int. Organ. Citrus Virol. Conf. Proc. 1996, 13, 267–270.
- Garnier, M.; Bové, J.M. Distribution of the huanglongbing (greening) Liberobacter species in fifteen African and Asian countries. Int. Organ. Citrus Virol. Conf. Proc. 1996, 13, 1957–2010.
- Bové, J.M.; Garnier, M. Citrus greening and psylla vectors of the disease in the Arabian Peninsula. Int. Organ. Citrus Virol. Conf. Proc. 1984, 9, 1957–2010.
- Promintara, M. Simultaneous infection of mandarin (Citrus reticulata Bl.) with tristeza virus and greening organism in Thailand. In Proceedings of the 4th International Asia Pacific Conference on Citrus Rehabilitation, Chiang Mai, Thailand, 4–10 February 1990; FAO-UNDP RAS/86/022 Regional Project. pp. 96–99.
- Bové, J.M.; Navarro, L.; Bonnet, P.; Zreik, L.; Garnier, M. Reaction of citrus cultivars to graft-inoculation of Phytoplasma aurantifolia-infected lime shoots. Int. Organ. Citrus Virol. Conf. Proc. (1957–2010) 1996, 13, 13.
- MAG. Declaraemergenciafitosanitarianacionalporpresencia de HLB, Enfermedad de Loscítricos, El Salvador, Ministerio de Agricultura y Ganadería. 2020. Available online: http://www.mag.gob.sv/mag-declara-emergencia-fitosanitaria-nacional-por-presencia-de-hlb-enfermedad-de-los-citricos/ (accessed on 25 November 2022).
- Cellier, G.; Moreau, A.; Cassam, N.; Hostachy, B.; Ryckewaert, P.; Aurela, L.; Picard, R.; Lombion, K.; Rioualec, A.L. First report of ‘Candidatus Liberibacter asiaticus’ associated with huanglongbing on Citrus latifolia in Martinique and Guadeloupe, French West Indies. Plant Dis. 2014, 98, 683.
- Leite, R., Jr.; Cordeiro, A.B.; Meneguim, L. Primera deteccion de la enfermedadHuanglongbing (HLB) enasociacion con la bacteria Candidatus Liberibacter asiaticus en Paraguay. In Proceedings of the VII Congresso Argentino de Citricultura, Puerto Iguazu, Missiones, Argentina, 15–17 May 2013.
- Marys, E.; Rodríguez-Román, E.; Mejías, R.; Mejías, A.; Mago, M.; Hernández, Y. First report on molecular evidence of Candidatus Liberibacter asiaticus associated with citrus Huanglongbing in Venezuela. J. Plant Pathol. 2020, 102, 1333.
- Aubert, B.; Garnier, M.; Cassin, J.E.; Bertin, Y. Citrus greening disease survey in East and West African countries South of Sahara. Int. Organ. Citrus Virol. Conf. Proc. (1957–2010) 1988, 10, 231–237.
- Garnier, M.; Jagoueix-Eveillard, S.; Cronje, P.R.; Le Roux, H.F.; Bove, J.M. Genomic characterization of a liberibacter present in an ornamental rutaceous tree, Calodendrumcapense, in the Western Cape Province of South Africa. Proposal of ‘Candidatus Liberibacter africanus subsp. capensis’. Int. J. Syst. Evol. Microbiol. 2000, 50, 2119–2125.
- Korsten, L.; Jagoueix, S.; Bové, J.M.; Garnier, M. Huanglongbing (greening) detection in South Africa. Int. Organ. Citrus Virol. Conf. Proc. (1957–2010) 1996, 13.
- Kalyebi, A.; Aisu, G.; Ramathani, I.; Ogwang, J.; McOwen, N.; Russell, P. Detection and identification of etiological agents (Liberibacter spp.) associated with citrus greening disease in Uganda. Uganda J. Agric. Sci. 2015, 16, 43–54.
- Ghosh, D.; Bhose, S.; Mukherjee, K.; Baranwal, V.K. Sequence and evolutionary analysis of ribosomal DNA from Huanglongbing (HLB) isolates of western India. Phytoparasitica 2013, 41, 295–305.
- Nageswara-Rao, M.; Miyata, S.I.; Ghosh, D.; Irey, M.; Garnsey, S.M.; Gowda, S. Development of rapid, sensitive and non-radioactive tissue-blot diagnostic method for the detection of citrus greening. Mol. Cell. Probes 2013, 27, 176–183.
- Hu, W.Z.; Wang, X.F.; Zhou, Y.; Li, Z.A.; Tang, K.Z.; Zhou, C. Diversity of the OMP gene in ‘Candidatus Liberibacter asiaticus’ in China. J. Plant Pathol. 2011, 93, 211–214.
- Kokane, S.B.; Bhose, S.; Kokane, A.; Gubyad, M.; Ghosh, D.K. Molecular detection, identification, and sequence analysis of Candidatus Liberibacter asiaticus associated with Huanglongbing disease of citrus in North India. 3Biotech 2020, 10, 341.
- Kokane, S.B.; Misra, P.; Kokane, A.D.; Gubyad, M.G.; Warghane, A.J.; Surwase, D.; Reddy, M.K.; Ghosh, D.K. Development of a real-time RT-PCR method for the detection of Citrus tristeza virus (CTV) and its implication in studying virus distribution in planta. 3Biotech 2021, 11, 431.
- Ghosh, D.K.; Kokane, A.; Kokane, S.B.; Tenzin, J.; Gubyad, M.G.; Wangdi, P.; Murkute, A.A.; Sharma, A.K.; Gowda, S. Detection and molecular characterization of Candidatus Liberibacter asiaticus and Citrus tristeza virus associated with citrus decline in Bhutan. Phytopathology 2021, 111, 870–881.
- Brlansky, R.H. Blight. In Compendium of Citrus Disease, 2nd ed.; Timmer, L.W., Garnsey, S.M., Graham, J.H., Eds.; APS Press: St. Paul, MN, USA, 2000; pp. 65–66.
- Tsai, J.H.; Liu, Y.H. Biology of Diaphorinacitri (Homoptera: Psyllidae) on four host plants. J. Econ. Entomol. 2000, 93, 1721–1725.
- Jagoueix, S.; Bové, J.M.; Garnier, M. The phloem-limited bacterium of greening disease of citrus is a member of the alpha subdivision of the proteobacteria. Int. J. Syst. Evol. Microbiol. 1994, 44, 379–386.
- Kokane, S.B.; Kokane, A.D.; Misra, P.; Warghane, A.; Kumar, P.; Gubyad, M.G.; Sharma, A.K.; Biswas, K.K.; Ghosh, D.K. In-silico characterization and RNA-binding protein based polyclonal antibodies production for detection of Citrus tristeza virus. Mol. Cell. Probes. 2020, 54, 101654.
- Ghosh, D.K.; Motghare, M.; Kokane, A.; Kokane, S.; Warghane, A.; Bhose, S.; Surwase, D.; Ladaniya, M.S. First report of a ‘Candidatus phytoplasma cynodontis’-related strain (group 16SrXIV) associated with Huanglongbing disease on Citrus grandis in India. Aus. Plant Dis. Notes 2019, 14, 9.
- Wang, N. A perspective of citrus Huanglongbing in the context of the mediterranean Basin. J. Plant Pathol. 2020, 102, 635–640.
- Pelz-Stelinski, K.S.; Brlansky, R.H.; Ebert, T.A.; Rogers, M.E. Transmission parameters for ‘Candidatus Liberibacter asiaticus’ by Asian citrus psyllid (Hemiptera: Psyllidae). J. Econ. Entomol. 2010, 103, 1531–1541.
- El-Shesheny, I.; Hijaz, F.; El-Hawary, I.; Mesbah, I.; Killiny, N. Impact of different temperatures on survival and energy metabolism in the Asian citrus psyllid, Diaphorina citri Kuwayama. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2016, 192, 28–37.
- Mann, R.S.; Pelz-Stelinski, K.; Hermann, S.L.; Tiwari, S.; Stelinski, L.L. Sexual transmission of a plant pathogenic bacterium, ‘Candidatus Liberibacter asiaticus’, between conspecific insect vectors during mating. PLoS ONE 2011, 6, e29197.
- Duan, Y.; Zhou, L.; Hall, D.G.; Li, W.; Doddapaneni, H.; Lin, H.; Liu, L.; Vahling, C.M.; Gabriel, D.W.; Kelly, P.; et al. Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant-Microbe Interact. 2009, 22, 1011–1020.
- Lin, H.; Han, C.S.; Liu, B.; Lou, B.; Bai, X.; Deng, C.; Civerolo, E.L.; Gupta, G. Complete genome sequence of a chinese strain of “Candidatus Liberibacter asiaticus. ” Genome Announc. 2013, 1, e00184-13.
- Katoh, H.; Miyata, S.; Inoue, H.; Iwanami, T. Unique features of a Japanese ‘Candidatus Liberibacter asiaticus’ strain revealed by whole genome sequencing. PLoS ONE 2014, 9, e106109.
- Zheng, Z.; Deng, X.; Chen, J. Whole-genome sequence of ‘Candidatus Liberibacter asiaticus’ from Guangdong, China. Genome Announc. 2014, 2, e00273-14.
- Zheng, Z.; Bao, M.; Wu, F.; Van Horn, C.; Chen, J.; Deng, X. A Type 3 Prophage of ‘Candidatus Liberibacter asiaticus’ carrying a restriction-modification system. Phytopathology 2018, 108, 454–461.
- Dai, Z.; Wu, F.; Zheng, Z.; Yokomi, R.; Kumagai, L.; Cai, W.; Rascoe, J.; Polek, M.; Chen, J.; Deng, X. Prophage diversity of ‘Candidatus Liberibacter asiaticus’ strains in california. Phytopathology 2019, 109, 551–559.
- Petrone, J.R.; Muñoz-Beristain, A.; Glusberger, P.R.; Russell, J.T.; Triplett, E.W. Unamplified, Long-Read Metagenomic Sequencing Approach to Close Endosymbiont Genomes of Low-Biomass Insect Populations. Microorganisms 2022, 10, 513.
- Li, S.; Wu, F.; Duan, Y.; Singerman, A.; Guan, Z. Citrus Greening: Management strategies and their economic impact. Hort. Sci. 2020, 55, 604.
- Wang, Y.; Kondo, T.; He, Y.; Zhou, Z.; Lu, J. Genome Sequence Resource of ‘Candidatus Liberibacter asiaticus’ from Diaphorinacitri Kuwayama (Hemiptera: Liviidae) in Colombia. Plant Dis. 2021, 105, 193–195.
- Lu, J.; Delatte, H.; Reynaud, B.; Beattie, G.A.; Holford, P.; Cen, Y.; Wang, Y. Genome sequence resource of ‘Candidatus Liberibacter asiaticus’ from Diaphorinacitri Kuwayama (Hemiptera: Liviidae) from La Réunion. Plant Dis. 2021, 105, 1171–1173.
- Toruno, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 2016, 54, 419–441.
- Thapa, S.P.; De Francesco, A.; Trinh, J.; Gurung, F.B.; Pang, Z.; Vidalakis, G.; Wang, N.; Ancona, V.; Ma, W.; Coaker, G. Genome-wide analyses of Liberibacter species provides insights into evolution, phylogenetic relationships, and virulence factors. Mol. Plant Pathol. 2020, 21, 716–731.
- Kruse, A.; Fleites, L.A.; Heck, M. Lessons from one fastidious bacterium to another: What can we learn about liberibacter species from Xylella fastidiosa. Insects 2019, 10, 300.
- Du, P.; Zhang, C.; Zou, X.; Zhu, Z.; Yan, H.; Wuriyanghan, H.; Li, W. “Candidatus Liberibacter asiaticus” secretes nonclassically secreted proteins that suppress host hypersensitive cell death and induce expression of plant pathogenesis-related proteins. Appl. Environ. Microbiol. 2021, 87, e00019-21.
- Pitino, M.; Armstrong, C.M.; Cano, L.M.; Duan, Y. Transient expression of Candidatus Liberibacter asiaticus effector induces cell death in Nicotiana benthamiana. Front. Plant Sci. 2016, 7, 982.
- Pitino, M.; Allen, V.; Duan, Y. LasΔ5315 effector induces extreme starch accumulation and chlorosis as Candidatus Liberibacter asiaticus infection in Nicotiana benthamiana. Front. Plant Sci. 2018, 9, 113.
- Singh, A.; Kumar, N.; Tomar, P.P.; Bhose, S.; Ghosh, D.K.; Roy, P.; Sharma, A.K. Characterization of a bacterioferritin comigratory protein family 1-Cys peroxiredoxin from Candidatus Liberibacter asiaticus. Protoplasma 2017, 254, 1675–1691.
- Wulff, N.A.; Zhang, S.; Setubal, J.C.; Almeida, N.F.; Martins, E.C.; Harakava, R.; Kumar, D.; Rangel, L.T.; Foissac, X.; Bové, J.M.; et al. The complete genome sequence of ‘Candidatus Liberibacter americanus’, associated with citrus huanglongbing. Mol. Plant Microbe Interact. 2014, 27, 163–176.
- Killiny, N. Quorum sensing controls Candidatus Liberibacter asiaticus interactions with host plant and insect vector. APS Annu. Meet. 2015, 105, 72.
- Shahzad, F.M.; Kadyampakeni, D.; Vashisth, T. Effect of Growing Media pH on Performance of Huanglongbing-Affected Young Citrus Trees. Agronomy 2021, 11, 439.
- Ramsey, J.S.; Chavez, J.D.; Johnson, R.; Hosseinzadeh, S.; Mahoney, J.E.; Mohr, J.P.; Robison, F.; Zhong, X.; Hall, D.G.; MacCoss, M.; et al. Protein interaction networks at the host-microbe interface in Diaphorinacitri, the insect vector of the citrus greening pathogen. Royal Soc. Open Sci. 2017, 4, 160545.
- Li, T.; Thaochan, N.; Huang, J.; Chen, J.; Deng, X.; Zheng, Z. Genome sequence resource of ‘Candidatus Liberibacter asiaticus’ from Thailand. Plant Dis. 2020, 104, 624–626.
- Zheng, Z.; Sun, X.; Deng, X.; Chen, J. Whole-genome sequence of “Candidatus Liberibacter asiaticus” from a huanglongbing-affected citrus tree in central Florida. Genome Announc. 2015, 3, e00169-15.
- Chen, Y.; Li, T.; Zheng, Z.; Xu, M.; Deng, X. Draft whole-genome sequence of a “Candidatus Liberibacter asiaticus” strain from Yunnan, China. Microbiol. Res. Announc. 2019, 8, e01413-18.
- Wu, F.; Zheng, Z.; Deng, X.; Cen, Y.; Liang, G.; Chen, J. Draft genome sequence of “Candidatus Liberibacter asiaticus” from Diaphorinacitri in Guangdong, China. Genome Announc. 2015, 3, e01316-15.
- Cai, W.; Nunziata, S.; Costanzo, S.; Kumagai, L.; Rascoe, J.; Stulberg, M.J. Genome resource for the huanglongbing causal agent ‘Candidatus Liberibacter asiaticus’ strain AHCA17 from citrus root tissue in California, USA. Plant Dis. 2020, 104, 627–629.
- Kunta, M.; Zheng, Z.; Wu, F.; da Graca, J.V.; Park, J.W.; Deng, X.; Chen, J. Draft whole-genome sequence of “Candidatus Liberibacter asiaticus” strain TX2351 isolated from Asian citrus psyllids in Texas, USA. Genome Announc. 2017, 5, e00170-17.
- Cai, W.; Yan, Z.; Rascoe, J.; Stulberg, M.J. Draft whole-genome sequence of “Candidatus Liberibacter asiaticus” strain TX1712 from citrus in texas. Genome Announc. 2018, 6, e00554-18.
- Liu, K.; Atta, S.; Cui, X.; Zeng, C.; Chen, J.; Zhou, C.; Wang, X. Genome sequence resources of two ‘Candidatus Liberibacter asiaticus’ strains from Pakistan. Plant Dis. 2020, 104, 2048–2050.
- Silva, P.A.; Huang, J.; Wulff, N.A.; Zheng, Z.; Krugner, R.; Chen, J. Genome sequence resource of ‘Candidatus Liberibacter asiaticus’ strain 9PA from Brazil. Plant Dis. 2021, 105, 199–201.
- Cui, X.J.; Zeng, C.H.; Liu, K.H.; Teng, C.L.; Zhou, C.Y.; Wang, X.F. Decreasing detection frequency of MITE (MCLas-A) in the population of ‘Candidatus Liberibacter asiaticus’ recently collected in southern China. J. Integr. Agric. 2020, 19, 2597–2601.
- Huang, J.; Dai, Z.; Zheng, Z.; Silvia, P. Metagenomic Analyses of ACP and Citrus Samples Infected with “Candidatus Liberibacter asiaticus” in California. In American Phytopathological Society Abstracts; USDA: Washington, DC, USA, 2020. Available online: https://www.ars.usda.gov/research/publications/publication/?seqNo115=373141 (accessed on 25 November 2022).
- Jain, M.; Fleites, L.; Gabriel, D.W. Prophage encoded peroxidase in “Candidatus Liberibacter asiaticus” is a secreted effector that suppresses plant defenses. Mol. Plant-Microbe Interac. 2015, 28, 1330–1337.
- Dominguez-Mirazo, M.; Jin, R.; Weitz, J.S. Functional and comparative genomic analysis of integrated prophage-like sequences in “Candidatus Liberibacter asiaticus”. mSphere 2019, 4, e00409-19.
- Zhang, S.; Flores-Cruz, Z.; Zhou, L.; Kang, B.H.; Fleites, L.A.; Gooch, M.D.; Wulff, N.A.; Davis, M.J.; Duan, Y.P.; Gabriel, D.W. ‘Ca. Liberibacter asiaticus’ carries an excision plasmid prophage and a chromosomally integrated prophage that becomes lytic in plant infections. Mol. Plant-Microbe Interact. 2011, 24, 458–468.
- Fu, S.M.; Hartung, J.; Zhou, C.Y.; Su, H.N.; Tan, J.; Li, Z.A. Ultrastructural changes and putative phage particles observed in sweet orange leaves infected with ‘Candidatus Liberibacter asiaticus’. Plant Dis. 2015, 99, 320–324.
- Zheng, Z.; Bao, M.; Wu, F.; Chen, J.; Deng, X. Predominance of single prophage carrying a CRISPR/cas system in “Candidatus Liberibacter asiaticus” strains in Southern China. PLoS ONE 2016, 11, e0146422.
- Castro, M.S.; Fontes, W. Plant defense and antimicrobial peptides. Protein Pept. Lett. 2005, 12, 11–16.
- Casteel, C.L.; Hansen, A.K.; Walling, L.L.; Paine, T.D. Manipulation of plant defense responses by the Tomato psyllid (Bactericercacockerelli) and its associated endosymbiont Candidatus Liberibacterpsyllaurous. PLoS ONE 2012, 7, e35191.
- Li, J.; Pang, Z.; Trivedi, P.; Zhou, X.; Ying, X.; Jia, H.; Wang, N. “Candidatus Liberibacter asiaticus” encodes a functional salicylic acid (SA) hydroxylase that degrades SA to suppress plant defenses. Mol. Plant-Microbe Interac. 2017, 30, 620–630.
- Clark, K.; Franco, J.Y.; Schwizer, S.; Pang, Z.Q.; Hawara, E.; Thomas, W.H.; Pagliaccia, D.; Zeng, L.; Gurung, F.B.; Wang, P.; et al. An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat. Commun. 2018, 9, 1718.
- Ying, X.; Wan, M.; Hu, L.; Zhang, J.; Li, H.; Dianqiu, L. Identification of the virulence factors of Candidatus Liberibacter asiaticus via heterologous expression in Nicotiana benthamiana using Tobacco mosaic virus. Int. J. Mol. Sci. 2019, 20, 5575.




