Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Seenivasan Ramasubbu | -- | 3409 | 2023-01-11 18:17:47 | | | |
| 2 | Rita Xu | Meta information modification | 3409 | 2023-01-12 03:59:46 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Florance, I.; Ramasubbu, S. Lipids in Macrophages of Different Tissue Location. Encyclopedia. Available online: https://encyclopedia.pub/entry/40070 (accessed on 08 February 2026).
Florance I, Ramasubbu S. Lipids in Macrophages of Different Tissue Location. Encyclopedia. Available at: https://encyclopedia.pub/entry/40070. Accessed February 08, 2026.
Florance, Ida, Seenivasan Ramasubbu. "Lipids in Macrophages of Different Tissue Location" Encyclopedia, https://encyclopedia.pub/entry/40070 (accessed February 08, 2026).
Florance, I., & Ramasubbu, S. (2023, January 11). Lipids in Macrophages of Different Tissue Location. In Encyclopedia. https://encyclopedia.pub/entry/40070
Florance, Ida and Seenivasan Ramasubbu. "Lipids in Macrophages of Different Tissue Location." Encyclopedia. Web. 11 January, 2023.
Copy Citation
Lipid metabolism is the major intracellular mechanism driving a variety of cellular functions such as energy storage, hormone regulation and cell division. Lipids, being a primary component of the cell membrane, play a pivotal role in the survival of macrophages. Lipids are crucial for a variety of macrophage functions including phagocytosis, energy balance and ageing. Lipid-loaded macrophages have recently been emerging as a hallmark for several diseases.
lipid droplets
macrophages
microglia
autophagy
hypoxia
1. Introduction
ER remains the key regulator of lipid metabolism in a cell. Around one-third of the proteome is synthesized, matured and modified at the rough ER, bound with ribosomes at the membrane. Biosynthesis of lipids, hormone, steroids and xenobiotic detoxification remains the function of smooth ER [1]. The ER houses the enzymes involved in synthesis of cholesterol and triacylglycerides (TAG) [2]. While TAGs are transferred to lipid droplets (LDs) budding from the ER membrane, lipids synthesized at ER are distributed to other organelles via the secretory pathway. At the ER, the cellular cholesterol is controlled via pathways that sense cholesterol levels within the ER membrane and impart signals to control both synthesis and clearance of cholesterol [3]. Under conditions of low ER cholesterol, the primary regulation of cholesterol at the ER involves synthesis of cholesterol via SCAP/SREBP2 (sterol regulatory-element binding proteins 2) pathway [4] followed by conversion of cholesterol into oxysterols and finally into bile acids [5] and production of cholesterol esters which eventually move into lipid droplets [6]. Similarly, ER regulates the intracellular fatty acid composition to regulate the cellular demands required for synthesizing complex lipids. Moreover, ER remains the key regulator of fatty acid synthesis and lipid metabolism. Structural modification of fatty acids such as elongases, desaturases and beta-oxidation cycles occur at ER [7]. SREBPs, a family of membrane bound transcription factors are actively involved in lipid homeostasis. SREBPs, synthesized as precursors reside at the ER membrane [8]. While SREBP1a functions mainly in lipid synthesis in proliferating cells, the major role of SREBP1c remains in regulating the synthesis of triglycerides (TG) and fatty acids in lipogenic organs. SREBP2 widely regulates synthesis of sterols in tissues [9][10]. De novo synthesis of fatty acids is also partially regulated at the ER through feedback inhibition mechanisms of SREBP-1 release [11]. Further, under stimulated TAG synthesis condition, enzymes involved in the biosynthesis of TAG (lipin, DGAT, GPAT and AGPAT) relocalize from ER to lipid droplets [12].
Additionally, synthesis of lipids occurs also at the ER-organelle contact sites. Lipids are trafficked out of ER by specialized ER domains via lipid transfer proteins [2]. It is scientifically plausible that lipid droplets are formed in ER at the regions where synthesis of triacylglycerols (TAGs) or sterol esters takes place [13]. Phospholipids and proteins are also biochemically modified at the ER–Golgi intermediate compartment (ERGIC) and are distributed within the cell via secretory pathways or direct organelle contacts [14][15]. Phospholipids and neutral lipids (TG and CE) are the two primary forms of lipids the ER is comprised of. The key function of phospholipids includes the assembly of membranes and vesicles involved in protein trafficking; TG and CE function as reserves for excess cholesterol and fatty acids and owing to their hydrophobicity, they instigate formation of LDs within the ER membrane [16]. Interestingly, ER -resident DGAT2 (diglyceride acyltransferase) enzyme, pivotal for the synthesis of TG, mediates synthesis and storage of TG in lipid droplets independent of its localization in ER [17]. Further, structurally uniform ER–LD contacts along with the delivery of TGs from ER to LDs were reported to be facilitated by an ER integral protein called seipin [18].
It is also notable that the PERK-eIF2α pathway regulates lipogenesis. For example, antipsychotic drugs induced phosphorylation of PERK and eIF2α, resulting in SREBP-1c and SREBP2 mediated accumulation of lipids in hepatic cells [19] (Figure 1).
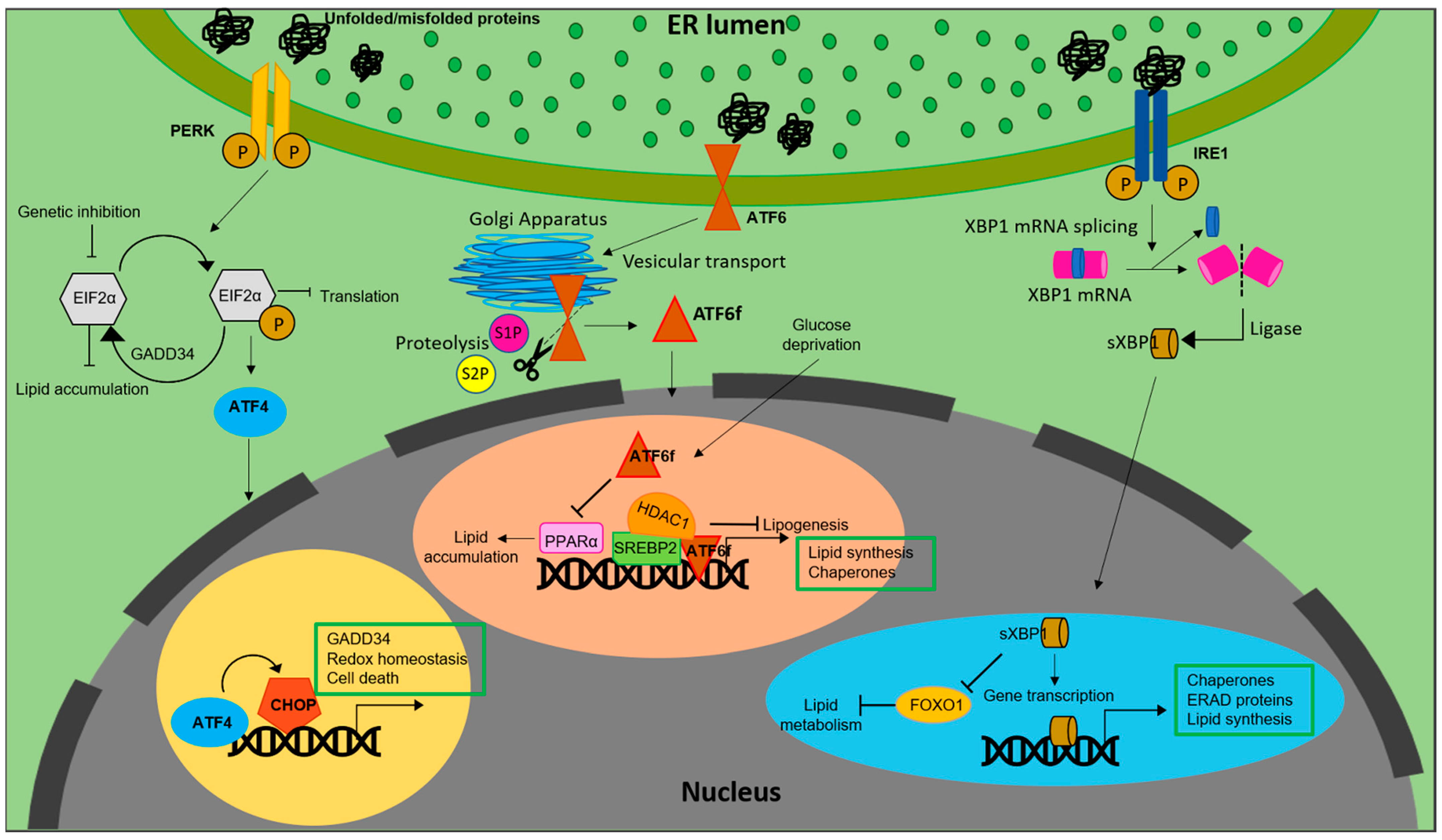
Figure 1. Lipid biosynthesis at ER. The schematic representation shows the mechanism of lipid biosynthesis taking place at the ER via the three branches of the UPR: ATF6, PERK and IRE1.
Similarly, inhibiting eIF2α phosphorylation via overexpression of growth arrest and DNA damage-inducible gene 34 (GADD34) in liver decreased the hepatosteatosis in mice fed with a high fat diet (HFD) [20]. Genetic deletion of eIF2α aggravated tunicamycin-induced accumulation of lipids in liver [21]. ER stress inducers, brefeldin A and tunicamycin, were reported to induce LD accumulation in Saccharomyces cerevisiae [22]. Further, ATF4, present downstream of the PERK-eIF2α pathway, plays a pivotal role in regulation of lipid metabolism. High carbohydrate diet fed Atf−/− mice displayed less accumulation of TG in liver compared to wild type [23]. Correspondingly, mice lacking ATF4 showed diminished lipid accumulation under conditions of high fructose diet due to decreased levels of FAS, acetyl CoA carboxylase (ACC) and SREBP-1c [24]. White adipose tissue of Atf deficient mice displayed increased lipolysis and decreased lipogenic genes, indicating a direct link between ATF4 and lipid metabolism [25]. Similarly, overexpressing ATF4 instigated early onset of dyslipidemia in zebrafish [26]. The involvement of (C/EBP) homologous protein (CHOP), a protein downstream of the UPR, in the regulation of lipid metabolism emanates from its role in suppressing the gene expression of SREBPF1, CEBPA, and PPARα-like master regulators of lipid metabolism [27]. Following ER stress, CHOP was reported to be critical for the regulation of cholesterol catabolism in macrophages and is involved in the lipid metabolism disorder mediated by ER stress [28]. In mammary epithelium and mouse embryonic fibroblasts that differentiate into adipocytes, absence of PERK resulted in the attenuation of lipogenesis and expression of genes including SREBP1. This led to a decrease in the level of TG and FA content in mammary glands and growth retardation in pups. The study also demonstrated the role of PERK and eIF2α in insig1 translation responsible for SREBP1 activation [29].
Another transmembrane signal transducer of the UPR, the inositol requiring enzyme 1 (IRE1α), is an ER stress sensor, conserved from yeast to mammals [30][31]. During ER stress, IRE1α is activated and splices X-box-binding protein 1 (XBP1) mRNA to its active spliced form to regulate expression of genes involved in restoration of ER homeostasis and biogenesis [32]. In addition to promoting cell survival through attenuation of ER stress, IRE1α also functions as a nutritional stress sensor [33]. Further, IRE1α was found to regulate lipid secretion and lipogenesis in both XBP1-dependent and an independent manner [34][35] (Figure 1). In c-Myc-overexpressing and IRE1α inhibited BL cells, defects in growth and viability were triggered due to altered lipid homeostasis [36]. The IRE1a/XBP1 signaling pathway transcriptionally regulates genes that are players of lipid metabolism in order to activate hepatic lipid metabolism. IRE1α siRNA increased TG and cholesterol levels in XBP1-deficient mice. This suggests that IRE1α hyperactivation reduced plasma lipids under XBP1-deficient conditions. Ablation of XBP1 decreased hepatoxicity [37]. However, how IRE1α functions to maintain lipid homeostasis in peripheral adipose tissues across species remains an enigma. As reviewed by Basseri et al., accumulating evidences suggest that IRE1α is the key component involved in the suppression of hepatic lipid accumulation under conditions of severe ER stress [38]. Under ER stress, IRE1α plays a pivotal role in hepatocytic secretion of LDL and VLDL [35]. XBP1s reduces lipid accumulation by promoting protein degradation of Forkhead box protein O1 (FOXO1) in cardiomyocytes. Similarly, overexpressing XBP1s specifically in cardiomyocytes, mitigated cardiac steatosis [39].
In contrast to IRE1α and PERK, activation of ATF6 does not involve phosphorylation. Under conditions of ER stress, ATF6 is released from BiP, after which the Golgi-localization sequences on the luminal domain of ATF6 are exposed [40] Once transported to Golgi, ATF6 undergoes site-1 (S1P) and site-2 (S2P) proteases mediated cleavage, releasing a cytosolic fragment called ATF6f containing a basic leucine zipper (bZIP) transcription factor [41][42]. The transcriptionally active ATF6 fragment enters the nucleus to trigger a set of transcriptional signaling, to reestablish the ER homeostasis [43][44][45]. Unlike PERK and IRE1 branches of UPR, ATF6 does not function to decrease the flux of unfolded proteins into the ER. Instead, the active fragment induces expansion of the ER membrane in an XBP1-independent manner [46] It also upregulates ER chaperones, ERAD components and disulfide oxidoreductases of the ER lumen [47]. However, ATF6 and XBP1 generated from the IRE1 branch of UPR tend to act synergistically and heterodimerize [48]. ATF6 activation downregulates PPARα, leading to accumulation of lipid droplets resulting in cell death. In contrast, deficiency of ATF6 increased PPARα levels and decreased lipid accumulation and cell death [49]. ATF6 was reported to modulate SREBP2 mediated lipogenesis. Under glucose-deprived conditions, ATF6 interacts with the processed form of SREBP2 to inhibit cholesterol synthesis promoting recruitment of HDAC1. This inhibits the SREBP2-induced lipogenesis and downregulates LDLR expression in HepG2 cells [50].
2. Functions of Lipids in Macrophages of Different Tissue Location
2.1. Lipids and Microglia
Microglia are specialized immune cells resident in the central nervous system (CNS) of the brain and play a key role in the maintenance of brain homeostasis [51][52][53]. Originating from the yolk sac, they migrate to the CNS during embryogenesis; there they propagate and disperse in the CNS in a non-heterogenous manner [54][55]. In a healthy resting brain, microglia are reported to be dynamic and constantly moving [56]. They critically survey the brain environment and get activated upon changes in the brain microenvironment [57]. Their functions include phagocytosis of apoptotic bodies and debris, neuronal protection [56], synaptic remodeling [58][59], neuronal support [60][61] and oligodendrogenesis [62][63]. Microglia dysfunction is a salient feature in neurodegenerative and neuroinflammatory diseases. They become dysfunctional with aging; dysfunctionalities include poor cholesterol efflux, impaired phagocytosis and increased secretion of cytokines and accumulation of lipid droplets [51][64]. Dysregulated lipid metabolism is a characteristic feature observed in neurodegenerative diseases, notably, in Parkinson’s disease (PD) and Alzheimer’s disease (AD). Lipid metabolism in microglia is tightly regulated both during development and disease. Microglia play an inevitable role in maintaining the myelin dynamics. During early development, microglia phagocytose myelin debris and apoptotic oligodendrocytes. The myelin-derived lipids are then cleared by microglia for remyelination post demyelination. Such active clearance and ability to effectively efflux cholesterol is impaired in aging microglia [65][66]. This leads to accumulation and crystallization of cholesterol-rich myelin debris, resulting in defective phagocytosis. Microglia with defective phagocytosis tend to produce large amounts of pro-inflammatory cytokines and reactive oxygen species (ROS) leading to the progression of neuroinflammatory and neurodegenerative diseases [67][68][69]. In a recent study, Loving et al., elucidated the role of lipoprotein lipase (LPL) in the accumulation of LDs and transcriptional regulation of lipid metabolism in in vitro and ex vivo systems. They reported that LPL regulates lipid metabolism in microglia and that loss of LPL resulted in microglial cholesterol load [64]. Phagocytosis deficit can be the consequence of lipid droplets accumulation. Active degradation of lipid droplets and release of free fatty acids was associated with effective phagocytosis [70].
Similarly, lipid droplet accumulating macrophages (LAMs) show downregulated expression of two key enzymes ADRB1 and ADRB2, involved in lipid degradation [51]. Additionally, lipid droplet accumulation promotes transcriptional modulation, giving LAMs a unique transcriptomic signature. Further, Patel and Tulsi et al. reported the association of pathways of lipid and carbohydrate metabolism with sex, age and ApoE expression in human microglia [71]. ApoE is a protein that mediates metabolism and transportation of cholesterol and is prominently expressed in disease-associated microglia (DAM). Moreover, it was reported that extracellular ApoE can be a ligand of TREM2 (triggering receptor expressed on myeloid cells 2) and that ApoE expression is TREM2-dependent [72][73]. Analysis of cell-specific lipidomics reveal that TREM2 deficiency mediates dysregulation of genes associated with lipid metabolism and leads to cholesterol ester overload in microglia [74]. Understanding the link between lipids and neuropathology of NPC (Niemann–Pick disease) patients revealed that loss of NPC1, an intracellular cholesterol transporter in microglia resulted in enhanced uptake of myelin but impaired myelin turnover. Macrophages derived from the blood of NPC patients were found to be similar to the pathological alterations exhibited by microglia of Npc1−/− mice. It was also revealed that Npc1−/− deficient microglia accumulated undigested lipid materials, indicating the role of Npc1 in lipid trafficking in microglia [75].
2.2. Lipids in Adipose Tissue Macrophages (ATMs)
Initially discovered for their role in microbial killing and phagocytosis, macrophages are now known to have distinct and context-dependent functions in different physiological settings. Lipids are the major source of energy for macrophages. Cell membranes of macrophages and precursors of bioactive lipids are provided by lipids. Lipids are also known to regulate the signal transduction during macrophage activation. Activation or polarization of macrophages are dependent on environmental stimuli, which are even tissue-specific, that dictate them to take up unique functions.
Adipose tissue macrophages (ATMs) are key players in metabolic diseases and obesity-associated inflammation. Circulating monocytes accumulating in adipose tissue lead to the development of ATMs [76]. In a study conducted by Prieur et al. in obese mice, it was observed that increase in the accumulation of lipids in ATMs resulted in polarization of macrophages into M1 phenotype, a phenotype associated with insulin resistance and obesity. Their results indicate that M1 polarization of ATMs are associated with accumulation and proliferation of lipid species, giving them the resemblance of vascular foam cells (discussed later). In addition to M1 polarization, ATMs of obese mice strongly accumulated lipids in their cytoplasm, resembling pro-atherosclerotic vascular foam cells. Increased fat deposition in adipose tissue decreased the expandability of adipose tissue, leading to adipocyte dysfunction and lipid leakage and thereafter lipid accumulation in ATMs [77]. Similarly, M1 macrophages treated with exogenous fatty acids showed an increase in TG and CE levels. The accumulation of exogenous fatty acids was high in M2 macrophages, revealing the impact of macrophage polarization on lipid composition and endogenous lipid pools [78]. ATMs of obese individuals crucially function to scavenge and eliminate adipocyte debris. Under increasing conditions of adiposity, excess lipid species are stored in ATMs, leading to the formation of lipid-laden ATM population [79]. Cd36, Fabp4, Fabp5 and Lpl are the set of genes that are highly conserved in lipid-associated ATMs [80].
Lipid-associated macrophages (LAMs) are distinctly conserved subset of macrophages predominantly expanded in adipose tissues of obese individuals. The formation of LAMs in adipose tissues are driven by a TF called TREM2. TREM2-regulated TAMs were reported to be inevitable during the loss of adipose tissue homeostasis, as they prevent metabolic disorders [80]. Suppression of tumor growth of triple-negative breast cancer (TNBC) was achieved by genetically depleting LAM subsets [81].
Atherosclerosis is another condition where the lipid homeostasis of macrophages is disrupted. During atherosclerosis, initially, the modified lipoproteins and serum lipids are deposited under the endothelial cells, activating them to secrete adhesion molecules. Circulating monocytes interact with these adhesion molecules that adhere to the endothelial cells and migrate to subendothelial space where they are differentiated into macrophages. Ingestion of modified lipoproteins by macrophages takes place via receptor mediated phagocytosis or pinocytosis. As a result of excess uptake of lipids, macrophages tend to store the excess neutral lipids in the form of lipid droplets in the cytoplasm. Excess accumulation of lipid droplets (LDs) in macrophages gives them a foamy appearance and hence they are called “macrophage foam cells”. These lipid-laden macrophages are the hallmark of atherosclerosis.
Formation of lipid-laden macrophage foam cells in lungs occur even during the infection of Mycobacterium tuberculosis. In response to TB infection, macrophages undergo metabolic changes and develop into foam cells. Though they resemble atherosclerotic foam cells, their lipid composition and roles remain different. Unlike atherosclerotic foam cells, their lipid content is predominantly triglycerides (TG) and not cholesterol. Here, the formation of TB foam cells is attributed to mycolic acid from pathogens. These TB foam cells often dominate the mycobacterial granulomata associated with caseum [82].
2.3. Lipids in Tumor Associated Macrophages (TAMs)
TAMs are macrophages present in the microenvironment of solid tumors, creating an immunosuppressive environment. Lipids play a key role in the development of TAMs in the tumor microenvironment (TME). Accumulating evidence suggest that abnormal lipid accumulation is inevitable for TAMs to engage in protumorigenic activity. Tumorigenesis and tumor progression is associated with the functional plasticity of TAMs which is often dictated by their metabolic features. The metabolic and functional landscape of tumor cells keeps evolving according to the selective pressure of the inconsistency in the availability of nutrients and oxygen in the TME, as a result of which functional features of TAMs are often altered [83]. Macrophages from both murine and human tumors were found to express high levels of a scavenger receptor, CD36, and ingest more lipids [84] for use as source of energy via oxidative phosphorylation and fatty acid oxidation. Contradictorily, enhanced fatty acid oxidation in macrophages caused by fatty acids in tumor microenvironment results in increased ROS production and decreased IL-10 secretion to eliminate tumor cells. This signifies the involvement of lipid metabolism in anti-tumor response [85]. Similarly, in prostatic adenocarcinoma (PCa), TAMs in the TME were shown to have dysregulated lipid metabolism. Accumulation of lipids in TAMs was reported to positively correlate with the progression of PCa [86]. TAMs characterized as M2-like cells, suppress tumor immune surveillance to promote tumor growth and metastasis. In a study, Wu et.al., demonstrated that enhancing lipid metabolism is sufficient for modulating the phenotype of macrophages into immunosuppressive TAMs [87]. Further, ingestion of a high amount of lipids from tumor cells leads to over expression of phosphoinositide 3-kinase (PI3K-γ) resulting in the polarization of TAMs into an M2-like phenotype. Inhibiting PI3K-γ reversed the pro-tumor phenotype of LD-loaded TAMs, suppressing the growth of gastric cancer [88]. In a study conducted on a melanoma model, it was reported that β-glucosylceramide released by tumor cells served as a stimulus for protumorigenic polarization of TAMs via induction of ER stress responses-mediated shuffling of lipid composition in macrophages [89]. In addition to lipid accumulation, TAMs exhibit decreased phagocytic activity with upregulated expression of programmed death ligand 1 (PD-L1); they block anti–tumor T cell responses to support immunosuppression [88]. From these reports, it is scientifically evident that, lipid droplets are critical cell structures that can be targeted for the development of a novel anti-tumor strategy and that reprogramming lipid metabolism can maximize the impact of anti-tumor therapies.
2.4. Lipids in Phagocytic Function of Macrophages
Macrophages are phagocytic in nature. Through phagocytosis, macrophages engulf foreign organisms and other invading pathogens, thereby defending the host against infection. The crosstalk between hypoxia and inflammation has a significant implication for infection and sterile inflammation in macrophages. Macrophages are the primary component of the innate immune response that is known for phagocytosis of invading pathogens and microorganism. Apoptotic cells are also eliminated by macrophages via phagocytosis. Formation of lipid-rich organelles, called lipid bodies or lipid droplets (LDs), occur in parallel with formation and maturation of phagosomes containing pathogens [90][91]. Infections with microbes such as bacteria, virus and other parasites induced LD accumulation in immune cells both clinically and experimentally [92][93]. These lipid bodies, formed in response to infections relocate within cytoplasm to interact with the phagosomes [94]. However, this association between LDs and phagosomes is yet ill-understood. Nevertheless, this interaction is accounted for the survival of pathogens within host cells.
2.5. Oxidized Phosphocholines in the Immune Function of Macrophages
Phosphocholines, belonging to the class of phospholipids, are a vital component of mammalian cells. Cell death occurring in the local environment of inflammatory or non-inflammatory tissue injury results in ROS generation. This can oxidize the phosphocholines present in the plasma membrane. Upon exposure to ROS, arachidonic acid-containing phospholipid 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine (PAPC), the membrane component of mammalian cells, is oxidized at different positions, creating a heterogenous mixture of lipids called oxPAPCs. The scavenger receptor CD36 present on macrophages, induce the uptake of oxPAPCs. This leads to formation of foam cells during atherosclerosis [95]. The internalization of oxPAPCs into endosomes and their transport to cytosol is also mediated by bacterial lipopolysaccharide (LPS) receptor, CD14 present on myeloid cells [96]. Post recognition by macrophages, oxPAPCs exert proinflammatory or anti-inflammatory activities based on the context in which they were encountered [97]. Pre-treatment with oxPAPCs modulated the phagocytosis of bacteria in macrophages [98]. In contrast, oxPLs trigger CD36-mediated phagocytosis of apoptotic cells in macrophages [99]. Further, oxPAPCs perform a metabolic rewiring in macrophages which is speculated to increase mitochondrial fitness. This, in turn, could prolong their lifespan [100]. In addition, pure lipids present in oxPAPCs contribute to metabolic hyperinflammation. In addition, oxPAPCs are stimulators of inflammation in airways. During lung injury, oxPAPCs promote secretion of IL-6 from alveolar macrophages [101]. In contrast, pre-treatment of macrophages with oxPAPCs was reported to block the response of nuclear factor-κB to LPS treatments. The competitive interaction of LPS and oxPAPCs with CD14 remains the underlying mechanism [102]. The mechanism of oxPAPCs mediated accumulation of lipids in macrophages is shown in Figure 2.
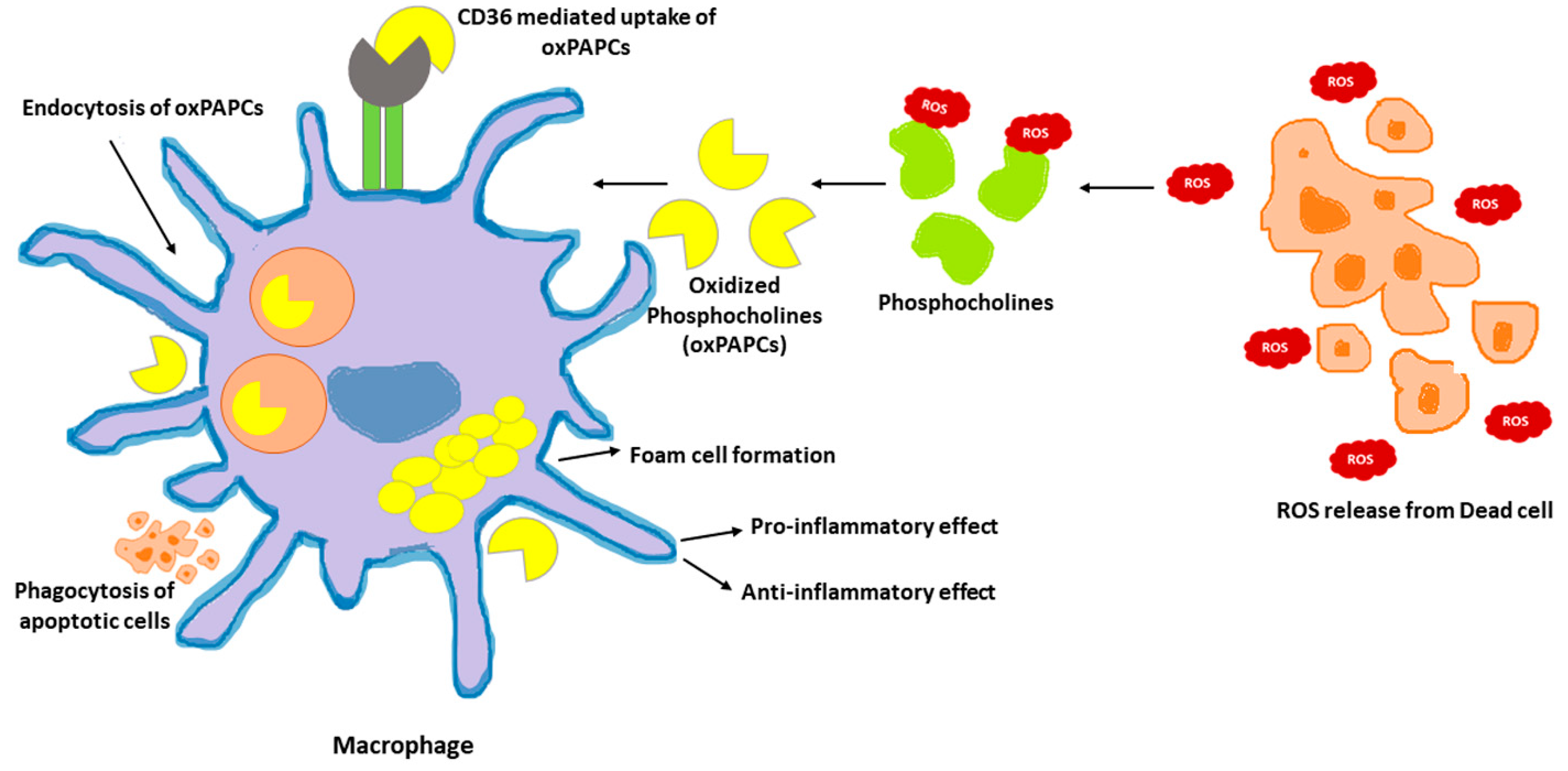
Figure 2. oxPAPCs in the regulation of macrophage immune function. Oxidized by the reactive oxygen speices (ROS) released from dead cells, phosphocholines are taken up by macrophages via endocytosis or CD36 scavenging receptors. Internalized oxPAPCs lead to the formation of foam cells by causing lipid accumulation in macrophages. The oxPAPCs accumulated macrophages are both pro- and anti-inflammatory based on the context.
References
- Xu, J.; Taubert, S. Beyond Proteostasis: Lipid Metabolism as a New Player in Er Homeostasis. Metabolites 2021, 11, 52.
- Jacquemyn, J.; Cascalho, A.; Goodchild, R.E. The Ins and Outs of Endoplasmic Reticulum-controlled Lipid Biosynthesis. EMBO Rep. 2017, 18, 1905–1921.
- Fu, S.; Watkins, S.M.; Hotamisligil, G.S. The Role of Endoplasmic Reticulum in Hepatic Lipid Homeostasis and Stress Signaling. Cell Metab. 2012, 15, 623–634.
- Brown, M.S.; Goldstein, J.L. Cholesterol Feedback: From Schoenheimer’s Bottle to Scap’s MELADL. J. Lipid Res. 2009, 50, S15–S27.
- Russell, D.W. The Enzymes, Regulation, and Genetics of Bile Acid Synthesis. Annu. Rev. Biochem. 2003, 72, 137–174.
- Martin, S.; Parton, R.G. Lipid Droplets: A Unified View of a Dynamic Organelle. Nat. Rev. Mol. Cell Biol. 2006, 7, 373–378.
- Miyazaki, M.; Ntambi, J.M. Fatty Acid Desaturation and Chain Elongation in Mammals. In Biochemistry of Lipids, Lipoproteins and Membranes; Elsevier: Amsterdam, The Netherlands, 2008; pp. 191–211. ISBN 9780444532190.
- Dorotea, D.; Koya, D.; Ha, H. Recent Insights Into SREBP as a Direct Mediator of Kidney Fibrosis via Lipid-Independent Pathways. Front. Pharmacol. 2020, 11, 265.
- Shimano, H.; Horton, J.D.; Shimomura, I.; Hammer, R.E.; Brown, M.S.; Goldstein, J.L. Isoform 1c of Sterol Regulatory Element Binding Protein Is Less Active than Isoform 1a in Livers of Transgenic Mice and in Cultured Cells. J. Clin. Investig. 1997, 99, 846–854.
- Horton, J.D.; Goldstein, J.L.; Brown, M.S. SREBPs: Activators of the Complete Program of Cholesterol and Fatty Acid Synthesis in the Liver. J. Clin. Investig. 2002, 109, 1125–1131.
- Horton, J.D. Sterol Regulatory Element-Binding Proteins: Transcriptional Activators of Lipid Synthesis. Biochem. Soc. Trans 2002, 30, 1091–1095.
- Wilfling, F.; Wang, H.; Haas, J.T.; Krahmer, N.; Gould, T.J.; Uchida, A.; Cheng, J.X.; Graham, M.; Christiano, R.; Fröhlich, F.; et al. Triacylglycerol Synthesis Enzymes Mediate Lipid Droplet Growth by Relocalizing from the ER to Lipid Droplets. Dev. Cell 2013, 24, 384–399.
- Zhang, P.; Reue, K. Lipin Proteins and Glycerolipid Metabolism: Roles at the ER Membrane and beyond. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1583–1595.
- Fagone, P.; Jackowski, S. Membrane Phospholipid Synthesis and Endoplasmic Reticulum Function. J. Lipid Res. 2009, 50.
- Appenzeller-Herzog, C.; Hauri, H.P. The ER-Golgi Intermediate Compartment (ERGIC): In Search of Its Identity and Function. J. Cell Sci. 2006, 119, 2173–2183.
- Farese, R.V.; Walther, T.C. Lipid Droplets Finally Get a Little R-E-S-P-E-C-T. Cell 2009, 139, 855–860.
- McFie, P.J.; Banman, S.L.; Kary, S.; Stone, S.J. Murine Diacylglycerol Acyltransferase-2 (DGAT2) Can Catalyze Triacylglycerol Synthesis and Promote Lipid Droplet Formation Independent of Its Localization to the Endoplasmic Reticulum. J. Biol. Chem. 2011, 286, 28235–28246.
- Salo, V.T.; Li, S.; Vihinen, H.; Hölttä-Vuori, M.; Szkalisity, A.; Horvath, P.; Belevich, I.; Peränen, J.; Thiele, C.; Somerharju, P.; et al. Seipin Facilitates Triglyceride Flow to Lipid Droplet and Counteracts Droplet Ripening via Endoplasmic Reticulum Contact. Dev. Cell 2019, 50, 478–493.e9.
- Han, J.; Kaufman, R.J. The Role of ER Stress in Lipid Metabolism and Lipotoxicity. J. Lipid Res. 2016, 57, 1329–1338.
- Oyadomari, S.; Harding, H.P.; Zhang, Y.; Oyadomari, M.; Ron, D. Dephosphorylation of Translation Initiation Factor 2α Enhances Glucose Tolerance and Attenuates Hepatosteatosis in Mice. Cell Metab. 2008, 7, 520–532.
- Rutkowski, D.T.; Wu, J.; Back, S.H.; Callaghan, M.U.; Ferris, S.P.; Iqbal, J.; Clark, R.; Miao, H.; Hassler, J.R.; Fornek, J.; et al. UPR Pathways Combine to Prevent Hepatic Steatosis Caused by ER Stress-Mediated Suppression of Transcriptional Master Regulators. Dev. Cell 2008, 15, 829–840.
- Fei, W.; Wang, H.; Bielby, C.; Yang, H. Conditions of Endoplasmic Reticulum Stress Stimulate Lipid Droplet Formation in Saccharomyces Cerevisiae. Biochem. J. 2009, 424, 61–67.
- Li, H.; Meng, Q.; Xiao, F.; Chen, S.; Du, Y.; Yu, J.; Wang, C.; Guo, F. ATF4 Deficiency Protects Mice from High-Carbohydrate-Diet-Induced Liver Steatosis. Biochem. J. 2011, 438, 283–289.
- Xiao, G.; Zhang, T.; Yu, S.; Lee, S.; Calabuig-Navarro, V.; Yamauchi, J.; Ringquist, S.; Dong, H.H. ATF4 Protein Deficiency Protects against High Fructose-Induced Hypertriglyceridemia in Mice. J. Biol. Chem. 2013, 288, 25350–25361.
- Wang, C.; Huang, Z.; Du, Y.; Cheng, Y.; Chen, S.; Guo, F. ATF4 Regulates Lipid Metabolism and Thermogenesis. Cell Res. 2010, 20, 174–184.
- Yeh, K.Y.; Lai, C.Y.; Lin, C.Y.; Hsu, C.C.; Lo, C.P.; Her, G.M. ATF4 Overexpression Induces Early Onset of Hyperlipidaemia and Hepatic Steatosis and Enhances Adipogenesis in Zebrafish. Sci. Rep. 2017, 7, 16362.
- Chikka, M.R.; McCabe, D.D.; Tyra, H.M.; Rutkowski, D.T. C/EBP Homologous Protein (CHOP) Contributes to Suppression of Metabolic Genes during Endoplasmic Reticulum Stress in the Liver. J. Biol. Chem. 2013, 288, 4405–4415.
- Sun, Y.; Zhang, D.; Liu, X.; Li, X.; Liu, F.; Yu, Y.; Jia, S.; Zhou, Y.; Zhao, Y. Endoplasmic Reticulum Stress Affects Lipid Metabolism in Atherosclerosis Via CHOP Activation and Over-Expression of miR-33. Cell. Physiol. Biochem. 2018, 48, 1995–2010.
- Bobrovnikova-Marjon, E.; Hatzivassiliou, G.; Grigoriadou, C.; Romero, M.; Cavener, D.R.; Thompson, C.B.; Diehl, J.A. PERK-Dependent Regulation of Lipogenesis during Mouse Mammary Gland Development and Adipocyte Differentiation. Proc. Natl. Acad. Sci. USA 2008, 105, 16314–16319.
- Walter, P.; Ron, D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science 2011, 334, 1081–1086.
- Zhao, P.; Huang, P.; Xu, T.; Xiang, X.; Sun, Y.; Liu, J.; Yan, C.; Wang, L.; Gao, J.; Cui, S.; et al. Fat Body Ire1 Regulates Lipid Homeostasis through the Xbp1s-FoxO Axis in Drosophila. iScience 2021, 24, 102819.
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA Is Induced by ATF6 and Spliced by IRE1 in Response to ER Stress to Produce a Highly Active Transcription Factor. Cell 2001, 107, 881–891.
- Huang, S.; Xing, Y.; Liu, Y. Emerging Roles for the ER Stress Sensor IRE1 in Metabolic Regulation and Disease. J. Biol. Chem. 2019, 294, 18726–18741.
- Lee, A.H.; Scapa, E.F.; Cohen, D.E.; Glimcher, L.H. Regulation of Hepatic Lipogenesis by the Transcription Factor XBP1. Science 2008, 320, 1492–1496.
- Zhang, K.; Wang, S.; Malhotra, J.; Hassler, J.R.; Back, S.H.; Wang, G.; Chang, L.; Xu, W.; Miao, H.; Leonardi, R.; et al. The Unfolded Protein Response Transducer IRE1Î ± Prevents ER Stress-Induced Hepatic Steatosis. EMBO J. 2011, 30, 1357–1375.
- Xie, H.; Tang, C.H.A.; Song, J.H.; Mancuso, A.; Del Valle, J.R.; Cao, J.; Xiang, Y.; Dang, C.V.; Lan, R.; Sanchez, D.J.; et al. IRE1α RNase-Dependent Lipid Homeostasis Promotes Survival in Myc-Transformed Cancers. J. Clin. Investig. 2018, 128, 1300–1316.
- So, J.S.; Hur, K.Y.; Tarrio, M.; Ruda, V.; Frank-Kamenetsky, M.; Fitzgerald, K.; Koteliansky, V.; Lichtman, A.H.; Iwawaki, T.; Glimcher, L.H.; et al. Silencing of Lipid Metabolism Genes through ire1α-Mediated Mrna Decay Lowers Plasma Lipids in Mice. Cell Metab. 2012, 16, 487–499.
- Basseri, S.; Austin, R.C. Endoplasmic Reticulum Stress and Lipid Metabolism: Mechanisms and Therapeutic Potential. Biochem. Res. Int. 2012, 2012, 1–13.
- Schiattarella, G.G.; Altamirano, F.; Kim, S.Y.; Tong, D.; Ferdous, A.; Piristine, H.; Dasgupta, S.; Wang, X.; French, K.M.; Villalobos, E.; et al. Xbp1s-FoxO1 Axis Governs Lipid Accumulation and Contractile Performance in Heart Failure with Preserved Ejection Fraction. Nat. Commun. 2021, 12, 1684.
- Shen, J.; Chen, X.; Hendershot, L.; Prywes, R. ER Stress Regulation of ATF6 Localization by Dissociation of BiP/GRP78 Binding and Unmasking of Golgi Localization Signals. Dev. Cell 2002, 3, 99–111.
- Haze, K.; Yoshida, H.; Yanagi, H.; Yura, T.; Mori, K. Mammalian Transcription Factor ATF6 Is Synthesized as a Transmembrane Protein and Activated by Proteolysis in Response to Endoplasmic Reticulum Stress. Mol. Biol. Cell 1999, 10, 3787–3799.
- Ye, J.; Rawson, R.B.; Komuro, R.; Chen, X.; Davé, U.P.; Prywes, R.; Brown, M.S.; Goldstein, J.L. ER Stress Induces Cleavage of Membrane-Bound ATF6 by the Same Proteases That Process SREBPs. Mol. Cell 2000, 6, 1355–1364.
- Wu, J.; Rutkowski, D.T.; Dubois, M.; Swathirajan, J.; Saunders, T.; Wang, J.; Song, B.; Yau, G.D.Y.; Kaufman, R.J. ATF6α Optimizes Long-Term Endoplasmic Reticulum Function to Protect Cells from Chronic Stress. Dev. Cell 2007, 13, 351–364.
- Shoulders, M.D.; Ryno, L.M.; Genereux, J.C.; Moresco, J.J.; Tu, P.G.; Wu, C.; Yates, J.R.; Su, A.I.; Kelly, J.W.; Wiseman, R.L. Stress-Independent Activation of XBP1s And/or ATF6 Reveals Three Functionally Diverse ER Proteostasis Environments. Cell Rep. 2013, 3, 1279–1292.
- Radanović, T.; Ernst, R. The Unfolded Protein Response as a Guardian of the Secretory Pathway. Cells 2021, 10, 2965.
- Bommiasamy, H.; Back, S.H.; Fagone, P.; Lee, K.; Meshinchi, S.; Vink, E.; Sriburi, R.; Frank, M.; Jackowski, S.; Kaufman, R.J.; et al. ATF6α Induces XBP1-Independent Expansion of the Endoplasmic Reticulum. J. Cell Sci. 2009, 122, 1626–1636.
- Maiuolo, J.; Bulotta, S.; Verderio, C.; Benfante, R.; Borgese, N. Selective Activation of the Transcription Factor ATF6 Mediates Endoplasmic Reticulum Proliferation Triggered by a Membrane Protein. Proc. Natl. Acad. Sci. USA 2011, 108, 7832–7837.
- Yamamoto, K.; Sato, T.; Matsui, T.; Sato, M.; Okada, T.; Yoshida, H.; Harada, A.; Mori, K. Transcriptional Induction of Mammalian ER Quality Control Proteins Is Mediated by Single or Combined Action of ATF6α and XBP1. Dev. Cell 2007, 13, 365–376.
- Moncan, M.; Mnich, K.; Blomme, A.; Almanza, A.; Samali, A.; Gorman, A.M. Regulation of Lipid Metabolism by the Unfolded Protein Response. J. Cell. Mol. Med. 2021, 25, 1359–1370.
- Zeng, L.; Lu, M.; Mori, K.; Luo, S.; Lee, A.S.; Zhu, Y.; Shyy, J.Y.J. ATF6 Modulates SREBP2-Mediated Lipogenesis. EMBO J. 2004, 23, 950–958.
- Marschallinger, J.; Iram, T.; Zardeneta, M.; Lee, S.E.; Lehallier, B.; Haney, M.S.; Pluvinage, J.V.; Mathur, V.; Hahn, O.; Morgens, D.W.; et al. Lipid-Droplet-Accumulating Microglia Represent a Dysfunctional and Proinflammatory State in the Aging Brain. Nat. Neurosci. 2020, 23, 194–208.
- Davalos, D.; Grutzendler, J.; Yang, G.; Kim, J.V.; Zuo, Y.; Jung, S.; Littman, D.R.; Dustin, M.L.; Gan, W.B. ATP Mediates Rapid Microglial Response to Local Brain Injury in Vivo. Nat. Neurosci. 2005, 8, 752–758.
- Nimmerjahn, A.; Kirchhoff, F.; Helmchen, F. Neuroscience: Resting Microglial Cells Are Highly Dynamic Surveillants of Brain Parenchyma in Vivo. Science 2005, 308, 1314–1318.
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Gokhan, S.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; et al. Fate Mapping Analysis Reveals That Adult Microglia Derive from Primitive Macrophages. Science 2010, 330, 841–845.
- Bachiller, S.; Jiménez-Ferrer, I.; Paulus, A.; Yang, Y.; Swanberg, M.; Deierborg, T.; Boza-Serrano, A. Microglia in Neurological Diseases: A Road Map to Brain-Disease Dependent-Inflammatory Response. Front. Cell. Neurosci. 2018, 12, 488.
- Wake, H.; Moorhouse, A.J.; Nabekura, J. Functions of Microglia in the Central Nervous System-beyond the Immune Response. Neuron Glia Biol. 2012, 7, 47–53.
- Hickman, S.E.; Kingery, N.D.; Ohsumi, T.K.; Borowsky, M.L.; Wang, L.C.; Means, T.K.; El Khoury, J. The Microglial Sensome Revealed by Direct RNA Sequencing. Nat. Neurosci. 2013, 16, 1896–1905.
- Paolicelli, R.C.; Bolasco, G.; Pagani, F.; Maggi, L.; Scianni, M.; Panzanelli, P.; Giustetto, M.; Ferreira, T.A.; Guiducci, E.; Dumas, L.; et al. Synaptic Pruning by Microglia Is Necessary for Normal Brain Development. Science 2011, 333, 1456–1458.
- Stephan, A.H.; Barres, B.A.; Stevens, B. The Complement System: An Unexpected Role in Synaptic Pruning during Development and Disease. Annu. Rev. Neurosci. 2012, 35, 369–389.
- Parkhurst, C.N.; Yang, G.; Ninan, I.; Savas, J.N.; Yates, J.R.; Lafaille, J.J.; Hempstead, B.L.; Littman, D.R.; Gan, W.B. Microglia Promote Learning-Dependent Synapse Formation through Brain-Derived Neurotrophic Factor. Cell 2013, 155, 1596–1609.
- Ueno, M.; Fujita, Y.; Tanaka, T.; Nakamura, Y.; Kikuta, J.; Ishii, M.; Yamashita, T. Layer v Cortical Neurons Require Microglial Support for Survival during Postnatal Development. Nat. Neurosci. 2013, 16, 543–551.
- Hagemeyer, N.; Hanft, K.M.; Akriditou, M.A.; Unger, N.; Park, E.S.; Stanley, E.R.; Staszewski, O.; Dimou, L.; Prinz, M. Microglia Contribute to Normal Myelinogenesis and to Oligodendrocyte Progenitor Maintenance during Adulthood. Acta Neuropathol. 2017, 134, 441–458.
- Wlodarczyk, A.; Holtman, I.R.; Krueger, M.; Yogev, N.; Bruttger, J.; Khorooshi, R.; Benmamar-Badel, A.; Boer-Bergsma, J.J.; Martin, N.A.; Karram, K.; et al. A Novel Microglial Subset Plays a Key Role in Myelinogenesis in Developing Brain. EMBO J. 2017, 36, 3292–3308.
- Loving, B.A.; Tang, M.; Neal, M.C.; Gorkhali, S.; Murphy, R.; Eckel, R.H.; Bruce, K.D. Lipoprotein Lipase Regulates Microglial Lipid Droplet Accumulation. Cells 2021, 10, 198.
- Bruce, K.D.; Gorkhali, S.; Given, K.; Coates, A.M.; Boyle, K.E.; Macklin, W.B.; Eckel, R.H. Lipoprotein Lipase Is a Feature of Alternatively-Activated Microglia and May Facilitate Lipid Uptake in the CNS during Demyelination. Front. Mol. Neurosci. 2018, 11, 57.
- Lloyd, A.F.; Miron, V.E. The pro-Remyelination Properties of Microglia in the Central Nervous System. Nat. Rev. Neurol. 2019, 15, 447–458.
- Cantuti-Castelvetri, L.; Fitzner, D.; Bosch-Queralt, M.; Weil, M.T.; Su, M.; Sen, P.; Ruhwedel, T.; Mitkovski, M.; Trendelenburg, G.; Lütjohann, D.; et al. Defective Cholesterol Clearance Limits Remyelination in the Aged Central Nervous System. Science 2018, 359, 684–688.
- Nadjar, A. Role of Metabolic Programming in the Modulation of Microglia Phagocytosis by Lipids. Prostaglandins Leukot. Essent. Fat. Acids 2018, 135, 63–73.
- Khatchadourian, A.; Bourque, S.D.; Richard, V.R.; Titorenko, V.I.; Maysinger, D. Dynamics and Regulation of Lipid Droplet Formation in Lipopolysaccharide (LPS)-Stimulated Microglia. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2012, 1821, 607–617.
- Chandak, P.G.; Radović, B.; Aflaki, E.; Kolb, D.; Buchebner, M.; Fröhlich, E.; Magnes, C.; Sinner, F.; Haemmerle, G.; Zechner, R.; et al. Efficient Phagocytosis Requires Triacylglycerol Hydrolysis by Adipose Triglyceride Lipase. J. Biol. Chem. 2010, 285, 20192–20201.
- Patel, T.; Carnwath, T.P.; Wang, X.; Allen, M.; Lincoln, S.J.; Lewis-Tuffin, L.J.; Quicksall, Z.S.; Lin, S.; Tutor-New, F.Q.; Ho, C.C.G.; et al. Transcriptional Landscape of Human Microglia Implicates Age, Sex, and APOE-Related Immunometabolic Pathway Perturbations. Aging Cell 2022, 21, e13606.
- Atagi, Y.; Liu, C.C.; Painter, M.M.; Chen, X.F.; Verbeeck, C.; Zheng, H.; Li, X.; Rademakers, R.; Kang, S.S.; Xu, H.; et al. Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2). J. Biol. Chem. 2015, 290, 26043–26050.
- Nugent, A.A.; Lin, K.; van Lengerich, B.; Lianoglou, S.; Przybyla, L.; Davis, S.S.; Llapashtica, C.; Wang, J.; Kim, D.J.; Xia, D.; et al. TREM2 Regulates Microglial Cholesterol Metabolism upon Chronic Phagocytic Challenge. Neuron 2020, 105, 837–854.e9.
- Damisah, E.C.; Rai, A.; Grutzendler, J. TREM2: Modulator of Lipid Metabolism in Microglia. Neuron 2020, 105, 759–761.
- Colombo, A.; Dinkel, L.; Müller, S.A.; Sebastian Monasor, L.; Schifferer, M.; Cantuti-Castelvetri, L.; König, J.; Vidatic, L.; Bremova-Ertl, T.; Lieberman, A.P.; et al. Loss of NPC1 Enhances Phagocytic Uptake and Impairs Lipid Trafficking in Microglia. Nat. Commun. 2021, 12, 1158.
- Hassnain Waqas, S.F.; Noble, A.; Hoang, A.C.; Ampem, G.; Popp, M.; Strauß, S.; Guille, M.; Röszer, T. Adipose Tissue Macrophages Develop from Bone Marrow–independent Progenitors in Xenopus Laevis and Mouse. J. Leukoc. Biol. 2017, 102, 845–855.
- Prieur, X.; Mok, C.Y.L.; Velagapudi, V.R.; Núñez, V.; Fuentes, L.; Montaner, D.; Ishikawa, K.; Camacho, A.; Barbarroja, N.; O’Rahilly, S.; et al. Differential Lipid Partitioning between Adipocytes and Tissue Macrophages Modulates Macrophage Lipotoxicity and M2/M1 Polarization in Obese Mice. Diabetes 2011, 60, 797–809.
- Morgan, P.K.; Huynh, K.; Pernes, G.; Miotto, P.M.; Mellett, N.A.; Giles, C.; Meikle, P.J.; Murphy, A.J.; Lancaster, G.I. Macrophage Polarization State Affects Lipid Composition and the Channeling of Exogenous Fatty Acids into Endogenous Lipid Pools. J. Biol. Chem. 2021, 297, 101341.
- Aouadi, M.; Vangala, P.; Yawe, J.C.; Tencerova, M.; Nicoloro, S.M.; Cohen, J.L.; Shen, Y.; Czech, M.P. Lipid Storage by Adipose Tissue Macrophages Regulates Systemic Glucose Tolerance. Am. J. Physiol. Endocrinol. Metab. 2014, 307, E374–E383.
- Jaitin, D.A.; Adlung, L.; Thaiss, C.A.; Weiner, A.; Li, B.; Descamps, H.; Lundgren, P.; Bleriot, C.; Liu, Z.; Deczkowska, A.; et al. Lipid-Associated Macrophages Control Metabolic Homeostasis in a Trem2-Dependent Manner. Cell 2019, 178, 686–698.e14.
- Timperi, E.; Gueguen, P.; Molgora, M.; Magagna, I.; Kieffer, Y.; Lopez-Lastra, S.; Sirven, P.; Baudrin, L.G.; Baulande, S.; Nicolas, A.; et al. Lipid-Associated Macrophages Are Induced by Cancer-Associated Fibroblasts and Mediate Immune Suppression in Breast Cancer. Cancer Res. 2022, 82, 3291–3306.
- Kim, M.J.; Wainwright, H.C.; Locketz, M.; Bekker, L.G.; Walther, G.B.; Dittrich, C.; Visser, A.; Wang, W.; Hsu, F.F.; Wiehart, U.; et al. Caseation of Human Tuberculosis Granulomas Correlates with Elevated Host Lipid Metabolism. EMBO Mol. Med. 2010, 2, 258–274.
- Zheng, X.; Mansouri, S.; Krager, A.; Grimminger, F.; Seeger, W.; Pullamsetti, S.S.; Wheelock, C.E.; Savai, R. Metabolism in Tumour-Associated Macrophages: A Quid pro Quo with the Tumour Microenvironment. Eur. Respir. Rev. 2020, 29, 200134.
- Su, P.; Wang, Q.; Bi, E.; Ma, X.; Liu, L.; Yang, M.; Qian, J.; Yi, Q. Enhanced Lipid Accumulation and Metabolism Are Required for the Differentiation and Activation of Tumor-Associated Macrophages. Cancer Res. 2020, 80, 1438–1450.
- Yu, W.; Lei, Q.; Yang, L.; Qin, G.; Liu, S.; Wang, D.; Ping, Y.; Zhang, Y. Contradictory Roles of Lipid Metabolism in Immune Response within the Tumor Microenvironment. J. Hematol. Oncol. 2021, 14.
- Masetti, M.; Carriero, R.; Portale, F.; Marelli, G.; Morina, N.; Pandini, M.; Iovino, M.; Partini, B.; Erreni, M.; Ponzetta, A.; et al. Lipid-Loaded Tumor-Associated Macrophages Sustain Tumor Growth and Invasiveness in Prostate Cancer. J. Exp. Med. 2021, 219, e20210564.
- Wu, H.; Han, Y.; Rodriguez Sillke, Y.; Deng, H.; Siddiqui, S.; Treese, C.; Schmidt, F.; Friedrich, M.; Keye, J.; Wan, J.; et al. Lipid Droplet-dependent Fatty Acid Metabolism Controls the Immune Suppressive Phenotype of Tumor-associated Macrophages. EMBO Mol. Med. 2019, 11, e10698.
- Luo, Q.; Zheng, N.; Jiang, L.; Wang, T.; Zhang, P.; Liu, Y.; Zheng, P.; Wang, W.; Xie, G.; Chen, L.; et al. Lipid Accumulation in Macrophages Confers Protumorigenic Polarization and Immunity in Gastric Cancer. Cancer Sci. 2020, 111, 4000–4011.
- Di Conza, G.; Tsai, C.H.; Gallart-Ayala, H.; Yu, Y.R.; Franco, F.; Zaffalon, L.; Xie, X.; Li, X.; Xiao, Z.; Raines, L.N.; et al. Tumor-Induced Reshuffling of Lipid Composition on the Endoplasmic Reticulum Membrane Sustains Macrophage Survival and pro-Tumorigenic Activity. Nat. Immunol. 2021, 22, 1403–1415.
- D’Avila, H.; Freire-de-Lima, C.G.; Roque, N.R.; Teixeira, L.; Barja-Fidalgo, C.; Silva, A.R.; Melo, R.C.N.; DosReis, G.A.; Castro-Faria-Neto, H.C.; Bozza, P.T. Host Cell Lipid Bodies Triggered by Trypanosoma Cruzi Infection and Enhanced by the Uptake of Apoptotic Cells Are Associated with Prostaglandin E2 Generation and Increased Parasite Growth. J. Infect. Dis. 2011, 204, 951–961.
- Melo, R.C.N.; Fabrino, D.L.; Dias, F.F.; Parreira, G.G. Lipid Bodies: Structural Markers of Inflammatory Macrophages in Innate Immunity. Inflamm. Res. 2006, 55, 342–348.
- D’Avila, H.; Melo, R.C.N.; Parreira, G.G.; Werneck-Barroso, E.; Castro-Faria-Neto, H.C.; Bozza, P.T. Mycobacterium Bovis Bacillus Calmette-Guérin Induces TLR2-Mediated Formation of Lipid Bodies: Intracellular Domains for Eicosanoid Synthesis In Vivo. J. Immunol. 2006, 176, 3087–3097.
- Peyron, P.; Vaubourgeix, J.; Poquet, Y.; Levillain, F.; Botanch, C.; Bardou, F.; Daffé, M.; Emile, J.F.; Marchou, B.; Cardona, P.J.; et al. Foamy Macrophages from Tuberculous Patients’ Granulomas Constitute a Nutrient-Rich Reservoir for M. Tuberculosis Persistence. PLoS Pathog. 2008, 4, e1000204.
- Cocchiaro, J.L.; Kumar, Y.; Fischer, E.R.; Hackstadt, T.; Valdivia, R.H. Cytoplasmic Lipid Droplets Are Translocated into the Lumen of the Chlamydia Trachomatis Parasitophorous Vacuole. Proc. Natl. Acad. Sci. USA 2008, 105, 9379–9384.
- Febbraio, M.; Hajjar, D.P.; Silverstein, R.L. CD36: A Class B Scavenger Receptor Involved in Angiogenesis, Atherosclerosis, Inflammation, and Lipid Metabolism. J. Clin. Investig. 2001, 108, 785–791.
- Zanoni, I.; Tan, Y.; Di Gioia, M.; Springstead, J.R.; Kagan, J.C. By Capturing Inflammatory Lipids Released from Dying Cells, the Receptor CD14 Induces Inflammasome-Dependent Phagocyte Hyperactivation. Immunity 2017, 47, 697–709.e3.
- Bochkov, V.; Gesslbauer, B.; Mauerhofer, C.; Philippova, M.; Erne, P.; Oskolkova, O.V. Pleiotropic Effects of Oxidized Phospholipids. Free Radic. Biol. Med. 2017, 111, 6–24.
- Knapp, S.; Matt, U.; Leitinger, N.; van der Poll, T. Oxidized Phospholipids Inhibit Phagocytosis and Impair Outcome in Gram-Negative Sepsis In Vivo. J. Immunol. 2007, 178, 993–1001.
- Greenberg, M.E.; Sun, M.; Zhang, R.; Febbraio, M.; Silverstein, R.; Hazen, S.L. Oxidized phosphatidylserine–CD36 Interactions Play an Essential Role in Macrophage-Dependent Phagocytosis of Apoptotic Cells. J. Cell Biol. 2006, 203, 2613–2625.
- Zhivaki, D.; Kagan, J.C. Innate Immune Detection of Lipid Oxidation as a Threat Assessment Strategy. Nat. Rev. Immunol. 2022, 22, 322–330.
- Di Gioia, M.; Zanoni, I. Dooming Phagocyte Responses: Inflammatory Effects of Endogenous Oxidized Phospholipids. Front. Endocrinol. (Lausanne) 2021, 12, 626842.
- Erridge, C.; Kennedy, S.; Spickett, C.M.; Webb, D.J. Oxidized Phospholipid Inhibition of Toll-Like Receptor (TLR) Signaling Is Restricted to TLR2 and TLR4: Roles for CD14, LPS-Binding Protein, and MD2 as Targets for Specificity of Inhibition. J. Biol. Chem. 2008, 283, 24748–24759.
More
Information
Subjects:
Cell Biology; Immunology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
12 Jan 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




