Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ayman El-Baz | -- | 2890 | 2022-12-28 18:03:51 | | | |
| 2 | Catherine Yang | Meta information modification | 2890 | 2022-12-29 01:53:04 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Fahmy, D.; Alksas, A.; Elnakib, A.; Mahmoud, A.; Kandil, H.; Khalil, A.; Ghazal, M.; Bogaert, E.V.; Contractor, S.; El-Baz, A. Artificial Intelligence and Radiomics Techniques. Encyclopedia. Available online: https://encyclopedia.pub/entry/39506 (accessed on 08 February 2026).
Fahmy D, Alksas A, Elnakib A, Mahmoud A, Kandil H, Khalil A, et al. Artificial Intelligence and Radiomics Techniques. Encyclopedia. Available at: https://encyclopedia.pub/entry/39506. Accessed February 08, 2026.
Fahmy, Dalia, Ahmed Alksas, Ahmed Elnakib, Ali Mahmoud, Heba Kandil, Ashraf Khalil, Mohammed Ghazal, Eric Van Bogaert, Sohail Contractor, Ayman El-Baz. "Artificial Intelligence and Radiomics Techniques" Encyclopedia, https://encyclopedia.pub/entry/39506 (accessed February 08, 2026).
Fahmy, D., Alksas, A., Elnakib, A., Mahmoud, A., Kandil, H., Khalil, A., Ghazal, M., Bogaert, E.V., Contractor, S., & El-Baz, A. (2022, December 28). Artificial Intelligence and Radiomics Techniques. In Encyclopedia. https://encyclopedia.pub/entry/39506
Fahmy, Dalia, et al. "Artificial Intelligence and Radiomics Techniques." Encyclopedia. Web. 28 December, 2022.
Copy Citation
Hepatocellular carcinoma (HCC) is the most common primary hepatic neoplasm. Thanks to recent advances in computed tomography (CT) and magnetic resonance imaging (MRI), there is potential to improve detection, segmentation, discrimination from HCC mimics, and monitoring of therapeutic response. Radiomics, artificial intelligence (AI), and derived tools have already been applied in other areas of diagnostic imaging with promising results.
deep learning
machine learning
AI
computed tomography
1. Artificial Intelligence (AI)
The term “artificial intelligence (AI)” refers to the computational capacity to carry out tasks that are comparable to those carried out by humans, with varying degrees of autonomy, to process unique raw data (inputs) and produce useful information (outputs) [1]. While the foundations of AI were laid decades ago, it was not until the advent of modern powerful computational technology, coupled with the ability to capture and store massive data quantities, that it became possible to realize AI capabilities in the tasks most important to radiology, such as pattern recognition/identification, preparation, object/sound recognition, problem-solving, disease prognostication, assessing the necessity of therapy, and providing patients and clinicians with prognostic data on treatment outcomes. Despite the fact that healthcare is a daunting area in which to apply AI, medical image analysis is becoming a very important application of this technology [2]. From the outset, it has been obvious that radiologists could benefit from the powerful capabilities of computers to augment the standard procedures used for disease detection and diagnosis. The use of computer-aided diagnosis (CAD) systems, which were forerunners to modern AI, has been encouraged to help radiologists detect and analyze potential lesions in order to distinguish between diseases, reduce errors, and increase efficiency [3]. As the nature of CAD systems is that they are tailored to a specific task, their variable reliability and the possibility of false positive results require that a qualified radiologist confirm CAD findings [3]. Consequently, continuous efforts are being made to improve the efficiency and accuracy of CAD and to promote the assistance that it can offer in routine clinical practice. The development of artificial neural networks (ANN) in the middle of the 20th century [4] and their subsequent progression, which has brought forth the concepts of machine learning (ML), deep learning (DL), and computational learning models, are the primary reasons for the rise of AI.
Machine learning (ML) is one of the most important applications of AI. The training process, in which computer systems adapt to input data during a training cycle [5], is the cornerstone of ML. Such models require large amounts of high-quality input data for training. The creation and use of large datasets structured in such a way that they can be fed into an ML model are sometimes referred to as “big data”. Through repeated training cycles, ML models can adapt and improve their accuracy in predicting correct data labels. When an appropriate level of accuracy is achieved, the model can be applied to new cases which were not a part of the training stages [6][7]. ML algorithms can be either supervised or unsupervised, depending on whether the input data are labeled by human experts or unlabeled and directly categorized by various computational methods [6][8]. An optimal ML model should include both the most important features needed to generate desired outputs and the most generic features that can be generalized to the general population, even though these features may not be defined in advance. Pattern recognition and image segmentation, in which different meaningful regions of a digital image (i.e., pixels or segments) are identified, are two common ML tasks in radiology. Both have been successfully used for a variety of clinical settings, diseases, and modalities [9][10][11].
Deep learning (DL) is a subtype of ML which uses multilayered artificial neural networks (ANNs) to derive high-level features from input data (similar to neuronal networks) and to perform complex tasks in medical imaging. Specifically, DL is useful in classification tasks and in automatic feature extraction, where it is able to solve the issues of partial detectability and feature accessibility when trying to extract information from these valuable data sources. The use of multilayered convolutional neural networks (CNNs) improves DL robustness by mimicking human neuronal networks in the training phase. If applied to unlabelled data, the automatic process of learning relies on image features being automatically clustered based on their natural variability. Due to the challenge of achieving completely unsupervised instruction, complex learning models are most commonly implemented with a degree of human supervision. The performance of CAD can be increased using ML and CNNs. CAD systems utilizing ML can be trained on a dataset from a representative population and then identify the features of a single lesion in order to classify it as normal or abnormal [12]. Algorithms based on statistics are the major focal point of both supervised and unsupervised learning [13]; however, there are important variations. Classification (i.e., categorizing an image as normal or abnormal based on labels provided in the training data) and regression (observing or finding new categories using inference on training sets) are the two main applications of supervised learning. Unlike supervised learning, unsupervised models use unlabelled/unclassified data. As a result, latent pattern recognition is accomplished through dimensionality reduction and feature clustering [13]. To determine the usefulness of this classification process, it must first be validated. The capacity to link basic diagnostic patterns and features of medical image modalities to a specific pathological and histological subtyping has led to the area of radiomics by merging DL-based image processing with clinical, and when suitable, pathological/histological data [14][15][16].
2. Radiomics
Radiomics is a recent translational field in which a variety of properties, including geometry, signal strength, and texture, are extracted from radiological images in order to record imaging patterns and categorize tumor subtypes or grades. Radiomics is typically utilized in systems with various variants for prognosis, monitoring, and determining how well a treatment is working [15][17]. “Images are more than pictures; they’re data.” The basic concept of radiomics is beautifully illustrated by Robert Gillies and colleagues’ intuitive and precise description [18]. Radiomics is classified into two types, namely, feature-based and DL-based radiomics, and is commonly used to analyze medical images at a low computational cost. While clinical evaluations are subject to inter-observer variability, results using radiomics are more accurate, stable, and reproducible, as automated radiomic characteristics are either generated statistically from ML-based complicated computational models during the training phases (DL-based) or computed using mathematical methods (feature-based). However, in order to achieve a correct diagnosis, the input data must be of high quality, with accurate labels (in the case of supervised learning) or a population-representative sample (in the case of unsupervised learning) [19].
2.1. Feature-Based Radiomics
In order to perform feature-based radiomics, a segmented volume of interest (VOI) for 3D data or region of interest (ROI) for 2D data is used. In order to avoid overfitting (an erroneous reliance on clinically irrelevant features), feature selection algorithms examine a subset of certain features after feature extraction and create robust prediction models. Overfitting usually occurs in datasets that are too homogeneous and lack enough representations of the target diseases. As a result, the chosen features may not be valid when applied to different data sets. Selecting features from heterogeneous and representative datasets decreases the likelihood of selecting narrowly applicable features and reduces the risk of overfitting. Feature-based radiomics does not necessitate large datasets, as the features are typically defined after only a short computation time. Notably, the majority of extracted features are very complex, and often do not correlate to recognizable pysiological or pathological features, limiting model interpretability. The feature-based radiomics process contains the key processing stages listed below.
Image pre-processing is a basic step in radiomics in which useful features are extracted. The primary goal of radiomics is the generation of quantitative features from radiology images [18][19][20][21][22][23]. The generated features and models should be both repeatable and general, particularly when using multi-variate multi-model data (i.e., with different scanners, modalities, and acquisition protocols), which is the case for most medical imaging centers. Several pre-processing steps are necessary to achieve these objectives. Correcting inhomogeneities in MRI images, reducing noise, spatial resampling, spatial smoothing, and intensity normalization are all common pre-processing steps for radiomic analyses [15][24][25].
Tumor segmentation is another extraction step in feature-based radiomics. Segmentation of MR or CT images for different types of tumors is typically performed manually, either in preparation for ML model development or in clinical practice for planning of radiotherapy or volumetric evaluation of treatment response [23]. It takes a great deal of effort to carry out 3D manual segmenting of lesions that have necrosis, contrast enhancement, and surrounding tissue. The contours have a direct effect on the radiomics analysis results, as the segmented ROIs define the input for the feature-based radiomics process. To handle this challenge, ML approaches are being developed for automatic tumor localization and segmentation [26][27].
Feature extraction from medical images may lead to several quantitative traits, the majority of which are tumor heterogeneity. Although many features can be extracted, features are typically divided into four subgroups:
-
Shape characteristics, such as sphericity, compactness, surface area, and maximum dimensions, reflect the geometric characteristics of the segmented VOI [23].
-
First-order statistical features or histogram-based features describe how the intensity signals of pixels/voxels are distributed over the segmented ROI/VOI. These features neglect the spatial orientation and spatial relationship between pixels/voxels [20].
-
Second-order statistical features or textural features are statistical relationships between the signal intensity of adjacent pixels/voxels or groups of pixels or voxels. These features serve to quantify intratumoral heterogeneity. Textural features are created by numerically characterizing matrices that encode the exact spatial connections between the pixels/voxels in the source image. The gray-level co-occurrence matrix (GLCM) [28] is the most widely used texture analysis matrix. The GLCM shows how many times two intensity levels appear in adjacent pixels or voxels within a given distance and in a defined direction. Multiple textural characteristics, including energy, contrast, correlation, variance, homogeneity, cluster prominence, dissimilarity, cluster inclination, and maximum likelihood, can be measured using the GLCM. The difference in intensity levels between one pixel/voxel and its 26-pixel 3D neighborhood is represented by the neighborhood gray-level different matrix (NGLDM). For each image intensity, the gray-level run length matrix (GLRLM) encodes the size of homogeneous runs [29]. Long-run emphasis (LRE), short-run emphasis (SRE), low gray-level run emphasis (LGRE), run percentage (RP), and high gray-level run emphasis (HGRE) can all be derived from the GLRLM. There are other matrices that capture pixel-wise spatial relationships and can be used to compute additional texture-based features [28].
-
Higher-order statistical features are quantified using statistical methods after applying complex mathematical transformations (filters), such as for pattern recognition, noise reduction, local binary patterns (LBP), histogram-oriented gradients, or edge enhancement. Minkowski functionals, fractal analysis, wavelet or Fourier transforms, and Laplacian transforms of Gaussian-filtered images (Laplacian-of-Gaussian) are examples of these mathematical transformations or filters [25].
Feature selection is a helpful step used to refine the set of extracted features. For implementation of image-based models for prediction and prognosis, the extracted quantitative features might not have equal significance. Redundancy, overly strong correlation, and feature ambiguity can cause data overfitting and increase image noise sensitivity in the dependent predictive models. Overfitting is a methodological error in which the developed model is overly reliant on features specific to the radiological data used in the training process (i.e., noise or image artifacts) rather than features of the disease in question. Overfitting results in a model with deceptively high classification scores on the training dataset and weak performance on previously unseen data. One way to reduce the risk of overfitting is to employ feature selection prior to the learning phase [5]. Supervised and unsupervised feature selection techniques are widely used in radiomics. Unsupervised feature selection algorithms disregard class labels in favor of eliminating redundant spatial features. Principal component analysis (PCA) and cluster analysis are widely used techniques for this type of feature selection [15]. Despite the fact that these approaches minimize the risk of overfitting, they seldom yield the best feature subset. Supervised feature selection strategies, on the other hand, consider the relations between features and class labels, which results in the selection of features based on how much they contribute to the classification task. In particular, supervised feature selection procedures select features that increase the discrimination degree between classes [23]. For supervised feature set reduction, there are three common methods:
-
Filter methods (univariate methods) examine how features and labels are related without taking into account their redundancy, or correlation. Minimum redundancy maximum significance, Student’s t-test, Chi-squared score, Fisher score, and the Wilcoxon rank sum test are among the most commonly used filter methods. While these feature selection methods are widely used, they do not take the associations and interactions between features into account [30][31].
-
Wrapper methods (multivariate methods), known as greedy algorithms, avoid the filter method constraint by looking at the entire space of features and considering the relationships between each feature and other features in the dataset. A predictive model is used to evaluate the output of a group of features. The consistency of a given technique’s output is used to test each new subset of features. Wrapper approaches are computationally intensive, as they strive to find the best-performing functional group of features. Forward feature selection, backward feature exclusion, exhaustive feature selection, and bidirectional search are all examples of wrapper methods [30][31].
-
Embedded approaches carry out the feature selection process as part of the ML model’s development; in other words, the best group of features is chosen in the model’s training phase. In this way, embedded approaches incorporate the benefits of both the filter and wrapper methods. Embedded approaches provide more reliability than filter methods, have a lower execution time than wrapper methods, and are not very susceptibility to data overfitting, as they take into account the interactions between features. The least absolute shrinkage and selection operator (LASSO), tree-based algorithms such as the random forest classifier, and ridge regression are examples of commonly used embedded methods [30][31].
Model generation and evaluation is the final step in feature-based radiomics. Following feature selection, a predictive model can be trained to predict a predetermined ground truth, such as tumor recurrence versus tissues changes related to treatment. The most commonly used algorithms in radiomics include the Cox proportional hazards model for censored survival data, support vector machines (SVM), neural networks, decision trees (such as random forests), logistic regression, and linear regression. To avoid overfitting of ML models when using supervised methods, datasets are usually split into training and validation subsets to ensure that these subsets maintain a sample distribution similar to the class distribution; this is particularly important for small or unbalanced datasets. After training and validating the model, a previously unseen testing subset of data is introduced to test the model. Optimally, the testing data should be similar to the actual data that the model will work on in real clinical settings, and should be derived from a different source (e.g., a different institution or instrument) than the training data. As a consequence, when testing a model’s performance, robustness, and reliability, the testing dataset is the gold standard. Alternatively, statistical approaches such as cross-validation and bootstrapping can be used for model output estimation without using an external testing dataset, particularly for small datasets.
2.2. DL-Based Radiomics
ANNs that mimic the role of human vision are used in DL-based radiomics to automatically generate higher-dimensional features from input radiological images at various abstraction and scaling levels. DL-based radiomics is particularly useful for pattern recognition and classification of high-dimensional and nonlinear data. The procedure is radically different from the one described above. In DL-based radiomics, medical images are usually analyzed using different network architectures or stacks of linear and nonlinear functions, such as auto-encoders and CNNs, to obtain the most significant characteristics. With no prior description or collection of features available, neural networks automatically identify those features of medical images which are important for classification [32]. Across the layers of a CNN, low-level features are combined to create higher-level abstract features. Finally, the derived features are used for analysis or classification tasks. Alternatively, the features derived from a CNN can be used to generate of other models, such as SVM, regression models, or decision trees, as is the case when using feature-based radiomic approaches. Feature selection is seldom used, because the networks produce and learn the critical features from the input data; instead, techniques such as regularization and dropout of learned link weights are used to avoid overfitting. Compared to feature-based radiomics, larger datasets are required in DL-based radiomics due to the high correlation between inputs and extracted features, which limits its applicability in many research fields that suffer from restrictions in data availability (such as neuro-oncological studies). Notably, the transfer learning approach can be used to circumvent this obstacle by employing pre-trained neural networks for a separate but closely related purpose [33]. Leveraging the prior learning of the network can achieve reliable performance even with limited data availability. These two types of radiomics and their different steps are illustrated in Figure 1.
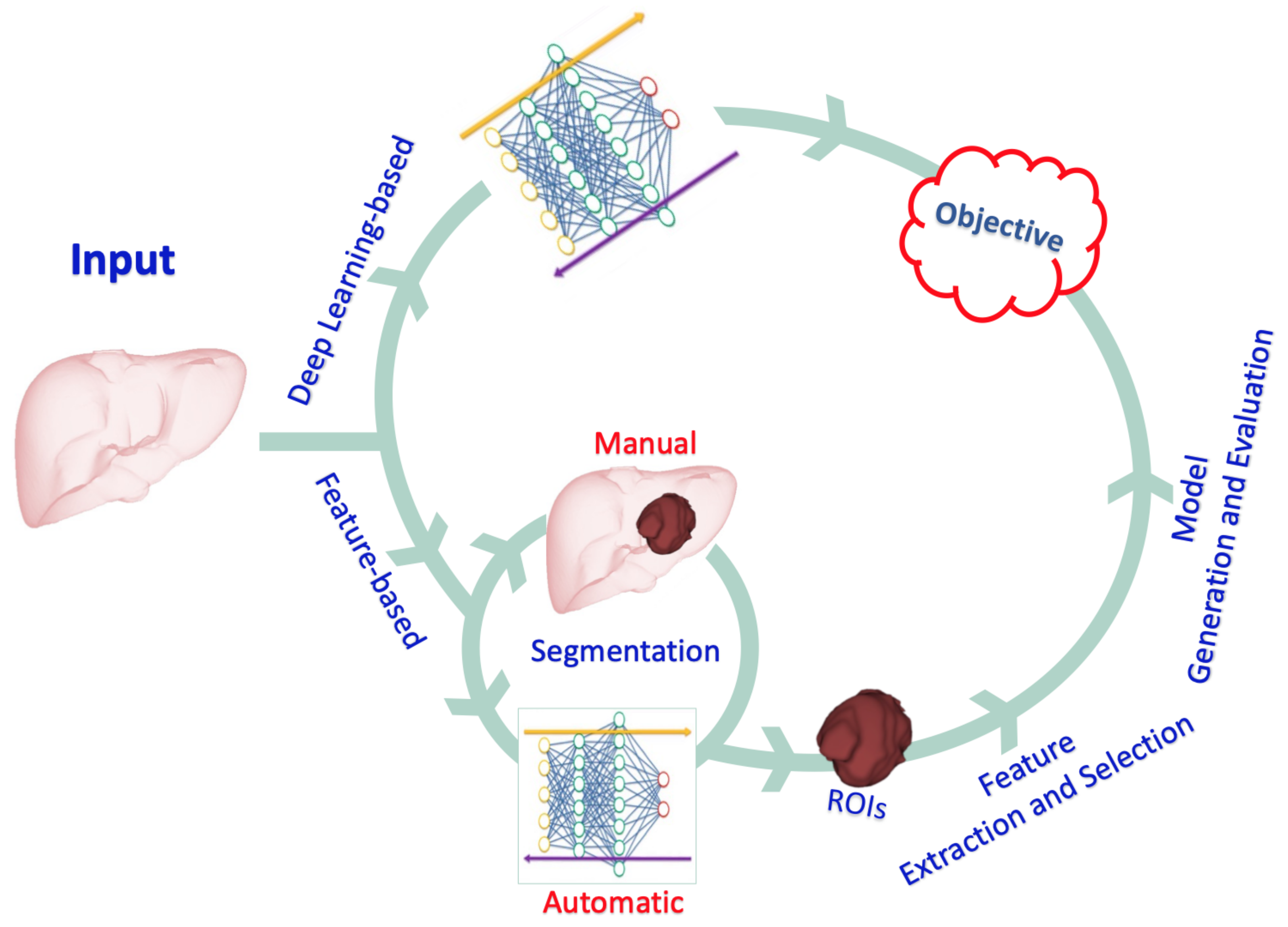
Figure 1. An illustration of the different types of radiomics and the steps involved.
3. Clinical Application of AI and Radiomic Techniques in Liver Cancer
Hepatocellular carcinoma (HCC), the most prevalent form of liver cancer, develops in the hepatocytes, the primary type of liver cell. Hepatoblastoma and intrahepatic cholangiocarcinoma are two significantly less frequent kinds of liver cancer. Because HCC is the most prevalent form of liver cancer, different AI and radiomics techniques can be used for HCC segmentation, detection, and management.
References
- Russell, S.; Norvig, P. Artificial Intelligence: A Modern Approach; Prentice Hall Press: Upper Saddle River, NJ, USA, 2003.
- Obermeyer, Z.; Emanuel, E.J. Predicting the future—Big data, machine learning, and clinical medicine. N. Engl. J. Med. 2016, 375, 1216.
- Castellino, R.A. Computer aided detection (CAD): An overview. Cancer Imaging 2005, 5, 17.
- Rosenblatt, F. The perceptron: A probabilistic model for information storage and organization in the brain. Psychol. Rev. 1958, 65, 386.
- SM, C.M. Artificial intelligence in radiology—Are we treating the image or the patient? Indian J. Radiol. Imaging 2018, 28, 137–139.
- Bishop, C.M.; Nasrabadi, N.M. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2006; Volume 4.
- Alpaydin, E. Introduction to Machine Learning; MIT Press: Cambridge, MA, USA, 2020.
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221.
- Saba, L.; Dey, N.; Ashour, A.S.; Samanta, S.; Nath, S.S.; Chakraborty, S.; Sanches, J.; Kumar, D.; Marinho, R.; Suri, J.S. Automated stratification of liver disease in ultrasound: An online accurate feature classification paradigm. Comput. Methods Programs Biomed. 2016, 130, 118–134.
- Saba, L.; Sanfilippo, R.; Tallapally, N.; Molinari, F.; Montisci, R.; Mallarini, G.; Suri, J.S. Evaluation of carotid wall thickness by using computed tomography and semiautomated ultrasonographic software. J. Vasc. Ultrasound 2011, 35, 136–142.
- Dey, D.; Gaur, S.; Ovrehus, K.A.; Slomka, P.J.; Betancur, J.; Goeller, M.; Hell, M.M.; Gransar, H.; Berman, D.S.; Achenbach, S.; et al. Integrated prediction of lesion-specific ischaemia from quantitative coronary CT angiography using machine learning: A multicentre study. Eur. Radiol. 2018, 28, 2655–2664.
- Kooi, T.; Litjens, G.; Van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312.
- Handelman, G.; Kok, H.; Chandra, R.; Razavi, A.; Lee, M.; Asadi, H. eD octor: Machine learning and the future of medicine. J. Intern. Med. 2018, 284, 603–619.
- Aerts, H.J. The potential of radiomic-based phenotyping in precision medicine: A review. JAMA Oncol. 2016, 2, 1636–1642.
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248.
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446.
- Abdel Razek, A.A.K.; Alksas, A.; Shehata, M.; AbdelKhalek, A.; Abdel Baky, K.; El-Baz, A.; Helmy, E. Clinical applications of artificial intelligence and radiomics in neuro-oncology imaging. Insights Imaging 2021, 12, 1–17.
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577.
- Mazurowski, M.A. Radiogenomics: What it is and why it is important. J. Am. Coll. Radiol. 2015, 12, 862–866.
- Lambin, P.; Leijenaar, R.T.; Deist, T.M.; Peerlings, J.; De Jong, E.E.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762.
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 1–9.
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 1–8.
- Alksas, A.; Shehata, M.; Saleh, G.A.; Shaffie, A.; Soliman, A.; Ghazal, M.; Khelifi, A.; Khalifeh, H.A.; Razek, A.A.; Giridharan, G.A.; et al. A novel computer-aided diagnostic system for accurate detection and grading of liver tumors. Sci. Rep. 2021, 11, 13148.
- Yip, S.S.; Aerts, H.J. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150.
- Wu, J.; Liu, A.; Cui, J.; Chen, A.; Song, Q.; Xie, L. Radiomics-based classification of hepatocellular carcinoma and hepatic haemangioma on precontrast magnetic resonance images. BMC Med. Imaging 2019, 19, 23.
- Huang, P.W.; Lai, Y.H. Effective segmentation and classification for HCC biopsy images. Pattern Recognit. 2010, 43, 1550–1563.
- Kim, D.W.; Lee, G.; Kim, S.Y.; Ahn, G.; Lee, J.G.; Lee, S.S.; Kim, K.W.; Park, S.H.; Lee, Y.J.; Kim, N. Deep learning–based algorithm to detect primary hepatic malignancy in multiphase CT of patients at high risk for HCC. Eur. Radiol. 2021, 31, 7047–7057.
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621.
- Xu, D.H.; Kurani, A.S.; Furst, J.D.; Raicu, D.S. Run-length encoding for volumetric texture. Heart 2004, 27, 452–458.
- Parekh, V.; Jacobs, M.A. Radiomics: A new application from established techniques. Expert Rev. Precis. Med. Drug Dev. 2016, 1, 207–226.
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013; Volume 26.
- Yasaka, K.; Akai, H.; Abe, O.; Kiryu, S. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: A preliminary study. Radiology 2018, 286, 887–896.
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A survey on deep transfer learning. In Proceedings of the International Conference on Artificial Neural Networks, Rhodes, Greece, 4–7 October 2018; Springer: New York, NY, USA, 2018; pp. 270–279.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
851
Revisions:
2 times
(View History)
Update Date:
30 Dec 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




